The skilled addressee will recognise tha be obtained by performing extra nested a 12-mer specificity could be obtained by performing two nested amplification reac |
t greater sequence specificity can mplification reactions. For example, designing three primer libraries and tions. |
EXAMPLE 1 |
PCR amplification of the fruR gene of E. |
coli. |
A 1727 bp fragment of the E. coli genome No. 1) was PCR amplified (FIG. 3). The P a 2 mM deoxynucleotide triphosphate mix triphosphates: 5 .mu.l of 10.times.PCR b (pH 9.0 at 25.degree. C.). 1% TritonX-10 1 .mu.l of the EK10FS primer (5'-tttaacc pmol per .mu.l); 1 .mu.l of the EK10R pr SEQ ID No. 3) (10 pmol per .mu.l); 1 .mu and 28 .mu.l of milliQ water. The PCR re C. in a PE 2400 PCR machine and 5 .mu.l unit of Taq DNA polymerase and 0.1 unit was subjected to 10 cycles of: 95.degree s reducing by 1.degree. C. per cycle, an reaction was immediately subjected to a for 10 s, 54.degree. C. for 20 s. 72.deg |
containing the fruR gene (SEQ ID CR reaction contained: 5 .mu.l of (dNTP) of all four deoxynucleotide uffer [500 mM KCl, 100 mM Tris-HCl 0]; 4 .mu.l of 25 mM MgCl.sub.2 ; cataccagtacaat-3': SEQ ID No. 2) (10 imer (5'-taacgggtaggcactgataag-3': .l of E. coli DNA (10 ng per .mu.l); action was preheated to 80.degree. of MilliQ water added containing 1 of Pfu DNA polymerase. The reaction . C. for 10 s, 65.degree. C. for 20 d 72.degree. C. for 90 s. The PCR further 20 cycles of: 95.degree. C. ree. C. for 90 s. |
ASIN PCR #1 |
Nested PCR amplifications of the 1727 bp using the ASIN procedure. First round AS .mu.l of 2 mM dNTPs: 1 .mu.l of 10.times : 1 .mu.l of the first round ASIN primer SEQ ID No. 4) (1 pmol per .mu.l); 0.2 .m DNA: 1 unit of Taq DNA polymerase: and 5 were preheated to 95.degree. C. for 30 s for 10 s. 40.degree. C. for 60 s. 72.deg |
E. coli fruR region were performed IN PCR amplifications contained: 1 .PCR buffer: 1 .mu.l of 25 mM MgCl.sub.2 TTCCTG (5'-cgattcgataacnnttcctg-3': u.l of the fruR PCR product as template .6 .mu.l of MilliQ water. PCR reactions before 2 cycles of: 95.degree. C. ree. C. for 60 s. |
ASIN PCR #2 |
The second round ASIN PCR mixes containe of 10.times.PCR buffer: 4 .mu.l of 25 mM second round ASIN primer (10 pmol per .m pmol per .mu.l): and 26 .mu.l of MilliQ ASIN primers were used: CTAG-TA (5'-gggc (FIG. 4: lane 1) and CTAG-GC (5'-gggcacg 4: lane 3). The second round PCR mixes w reactions and subjected to 25 cycles of: C. for 20 s. 72.degree. C. for 60 s. |
d: 4 .mu.l of 2 mM dNTPs: 4 .mu.l MgCl.sub.2 ; 1 .mu.l of the desired u.l); 1 .mu.l of EK10R primer (10 water. The following second round acgattcgataacta-3': SEQ ID No. 5) attcgataacgc-3': SEQ ID No. 6) (FIG. ere added to the first round ASIN 95.degree. C. for 10 s. 50.degree. |
6 .mu.l of each ASIN PCR amplification w buffer (40 mM Tris-Acetate, 1 mM EDTA) a with ethidium before being photographed |
ere run on a 2% agarose gel in 1.times.TAE t 40V for 1 h. The gel was stained under UV transillumination. |
EXAMPLE 2 |
ASIN PCR #1 |
PCR amplifications of region of the E. c on the pUC19 plasmid (plasmid pUC4G; J. using the ASIN procedure. First round AS .mu.l of 2 mM dNTPs: 2.5 .mu.l of 10.tim Tris-HCl (pH 9.0 at 25.degree. C.), 1% T ; 1 .mu.l of either of the two forward f tcgataacnnnctgcaa-3': SEQ ID No. 8) or 4 ID No. 9) (10 pmol per .mu.l): 1 .mu.l o 4G6 (5'-gattcgataacnnntgacca-3': SEQ ID of the pUC4G template DNA (FIG. 5) (5 ng |
oli, fusA gene (SEQ ID No. 7) cloned Bacteriol. 176: 123-29) were performed IN PCR amplifications contained: 2.5 es.PCR buffer [500 mM KCl, 100 mM ritonX-100]; 2.5 .mu.l of 25 mM MgCl.sub.2 irst round ASIN primers, 4G1 (5'-gat G3 (5'-gattcgataacnnntgagcg-3': SEQ f the reverse first round ASIN primer No. 10) (10 pmol per .mu.l): 1 .mu.l per .mu.l). |
The PCR reactions were preheated to 80.d and 5 .mu.l of MilliQ water added contai The reaction was subjected to 5 cycles o C. for 1 min, 72.degree. C. for 2 min. T ted to a further 10 cycles of: 95.degree s. 72.degree. C. for 2 min. |
egree. C. in a PE 2400 PCR machine ning 1 unit of Taq DNA polymerase. f: 95.degree. C. for 10 s, 39.degree. he PCR reaction was immediately subjec- . C. for 10 s, 54.degree. C. for 30 |
Control ASIN PCR #1 reactions were perfo the PCR conditions were modified so that C. for 10 s. 54.degree. C. for 30 s, 72. to 25 cycles of: 95.degree. C. for 10 s. C. for 2 min (FIG. 6: lanes 2 & 4). |
rmed as described previously except the final 10 cycles of: 95.degree. degree. C. for 2 min were changed 54.degree. C. for 30 s, 72.degree. |
ASIN PCR #2 |
The second round ASIN PCR mixes containe of 10.times.PCR buffer: 2.5 .mu.l of 25 second round ASIN primer (10 pmol per .m The following second round ASIN primers ac-3': SEQ ID No. 11) and 4G9 (5'-ggcacg the 4G3/4G6 ASIN PCR #1 reaction (FIG. 6 ac-3') and 4G7 (5'-ggcacgattcgataacgcg-3 ASIN PCR #1 (FIG. 6; lane 5). 1 .mu.l of was added to the second round ASIN react |
d: 2.5 .mu.l of 2 mM dNTPs: 2.5 .mu.l mM MgCl.sub.2 : 1 .mu.l of the desired u.l); and 10.5 .mu.l of MilliQ water. were used: 4G12 (5'-gggcacgattcgataact- attcgataacacg-3': SEQ ID No. 12) with : lane 3); 4G12 (5'-gggcacgattcgataact- ': SEQ ID No. 13) with the 4G1/4G6 the appropriate ASIN PCR #1 product ions as template DNA. |
The PCR reaction was preheated to 80.deg 5 .mu.l of MilliQ water added containing reaction was subjected to 28 cycles of: C. for 30 s. 72.degree. C. for 2 min. |
ree. C. in a PE 2400 PCR machine and 1 unit of Taq DNA polymerase. The 95.degree. C. for 10 s. 47.degree. |
5 .mu.l of each ASIN PCR amplification w buffer (40 mM Tris-Acetate, 1 mM EDTA) a with ethidium bromide before being photo EXAMPLE 3 |
ere run on a 2% agarose gel in 1.times.TAE t 70V for 40 min. The gel was stained graphed under UV transillumination. |
The amplification of the target DNA usin tide sequences of ASIN 1st and ASIN 2nd were shown in Table 1. |
g lambda DNA as a template. The nucleo- oligomers to amplify the target DNAs |
TABLE 1 |
Target |
No. ASIN 1st oliogomer AS |
IN 2nd oliogomer |
1 CACACAGGAAACAGCTATGACNNNCGCTAC |
CACACAGGAAACAGCTATGACCTT |
(SEQ ID NO. 14) (SEQ |
ID NO. 17) |
2 CACACAGGAAACAGCTATGACNNNCGATTT |
CACACAGGAAACAGCTATGACGGT |
(SEQ ID NO. 15) (SEQ |
ID NO. 18) |
CACACAGGAAACAGCTATGACNNNAACGCA |
CACACAGGAAACAGCTATGACGGT |
(SEQ ID NO. 16) (SEQ |
ID NO. 19) |
The first reaction was carried out in 10 Taq buffer (Takara Shuzo Co. Ltd.), 0.2 Shuzo Co. Ltd), one pmol of ASIN 1st oli ase (Takara Shuzo Co. Ltd.), for the fir 30 seconds followed by two cycles of 95. C. for one minute; and 72.degree. C. for PCR mixture containing Taq buffer, 0.2 m and 10 pmol of reverse primer (GTCGATAAA added to the reaction solution. PCR was C. for ten seconds; 55.degree. C. for on minutes. The amplified product was furth akara Shuzo Co. Ltd.) to remove excess p purified product was determined by direc (CAGGAAACAGCTATGAC: SEQ ID No. 21). With No.3, 0.6 kb. 0.4 kb, and 1.7 kb of ampl ely, and the identified nucleotide seque imately 0.4 kb, was identical to that of by Sanger et al. in Journal of Molecular |
.mu.L of reaction mixture containing mM of dNTP, 2.5 ng of lambda DNA (Takara gomer, and one unit of Taq DNA polymer- st one cycle at 95.degree. C. for degree. C. for ten seconds: 35.degree. three minutes. Next, 40 .mu.L of M of dNTP, 10 pmol of ASIN 2nd oligomer, TGGGCAATACGAAC: SEQ ID No. 20) was performed for 30 cycles of 95.degree. e minute; and 72.degree. C. for three er purified by using Microcon YM-100(T- rimer. The nucleotide sequence of t cycle sequencing using M13 RV primer respect to target No.1, No.2, and ified products were obtained respectiv- nce of these amplified products approx- target DNAs, which had been reported Biology, 162, 729-773 (1982). |
EXAMPLE 4 |
ASIN PCR#1 |
PCR amplifications of region of the E. c plasmid (plasmid pUC4G; J. Bacteriol. 17 ASIN procedure. First round ASIN PCR amp 2 mM dNTPs; 2.5 .mu.l of 10.times. PCR b (pH 9.0 at 25.degree. C.), 1% TritonX-10 ; 1 .mu.l of either of the two forward f gataacnnnetgeaa-3') or 4G3 (5'-gattegata 1 .mu.l of the reverse first round ASIN (10 pmol per .mu.l);1 .mu.l of the pUC4G |
oli fusA gene cloned on the pUC19 6: 123-129) were performed using the lifications contained; 2.5 .mu.l of uffer [500 mM KCl, 100 mM Tris-HCl 0]; 2.5 .mu.l of 25 mM MgCl.sub.2 irst round ASIN primers, 4G1 (5'-gatte- acnnntgageg-3') (10 pmol per .mu.l); primer 4G6 (5'-gattegataacnnntgacca-3') template DNA (5 ng per .mu.l). |
The PCR reactions were preheated to 80.d and 5 .mu.l of MilliQ water added contai The reaction was subjected to 5 cycles o C. for 1 min. 72.degree. C. for 2 min. T ted to a further 25 cycles of: 95.degree s. 72.degree. C. for 2 min. |
egree. C. in a PE 2400 PCR machine ning 1 unit of Taq DNA polymerase. f: 95.degree. C. for 10 s, 39.degree. he PCR reaction was immediately subjec- . C. for 10 s. 54.degree. C. for 30 |
DNA sequencing
|
DNA sequencing reactions were performed
resulting from 4G1/4G6 (T1) and 4G3/4G6
was removed from T1 and T2 by ethanol pr
product 2 .mu.l of 3 M sodium acetate (p
was added. The samples were incubated at
centrifugation at 12000 g for 5 min. The
DNA pellets which were allowed to air dr
of water.
|
using the mixed template products (T2) amplifications. Excess primer ecipitation. To 20 .mu.l of each PCR H 5.2) and 22 .mu.l of 85% ethanol room temperature for 5 min before supernatant was removed from the y for 30 min and dissolved in 8 .mu.l |
DNA sequencing reactions were performed
RR mix (ABI, Foster City, Calif.); 0.5 .
cgeg-3') with template T1, or 4G9 (5'-gg
T2: 3 .mu.l of either T1 or T2 DNA templ
The sequencing reactions were subjected
10 s. 45.degree. C. for 20 s. and 60.deg
reactions were purified using n-butanol
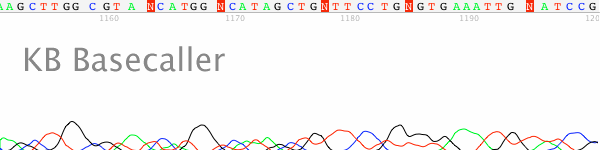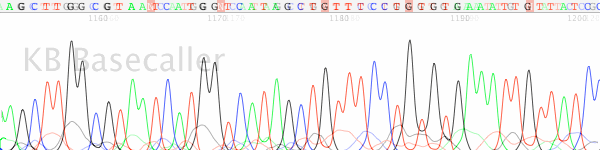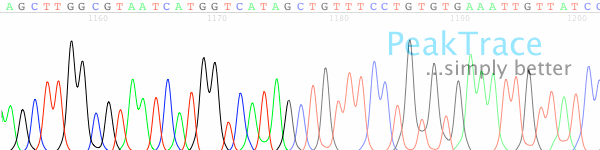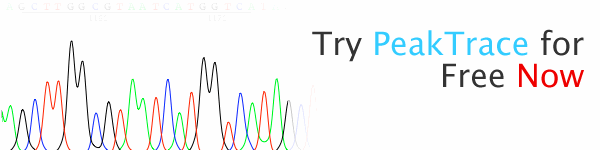
analysis on ABI 377 DNA sequencer. The r
that previously obtained from the pUC4G
|
using: 4 .mu.l of BigDye Terminator mu.l of either 4G7 (5'-ggcacgattegataa- cacgattegataacacg-3') with template ates: and 2.5 .mu.l of milliQ water. to 25 cycles of: 95.degree. C. for ree. C. for 4 min. The sequencing (Biotechniques 26; 606-610) before esulting DNA sequence data matched plasmid. |
50. A method according to claim 49 where sequence complementary to the first know of interest is 3 bases in length. |
in, in the sequencing primer, the n region of the nucleotide sequence |