Identification of DNA Sequence Slippages
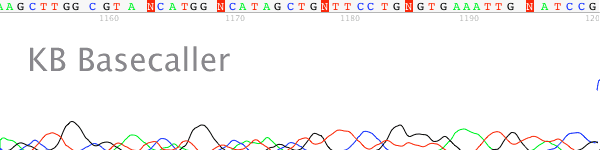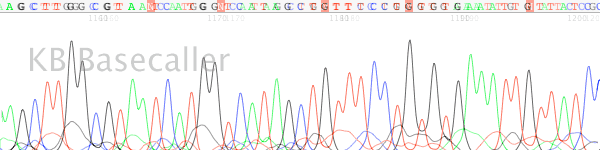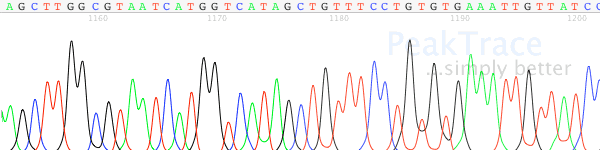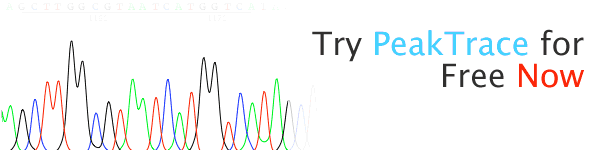
- The trace data becomes mixed after a long mononucleotide (single base) run (most typically A or T) in the DNA template (Figures 1 & 2).
- Can also occur on long dinucleotide AT repeats in a manner analogous to stutter bands that occur in microsatellite polymerase chain reaction (PCR) amplification.
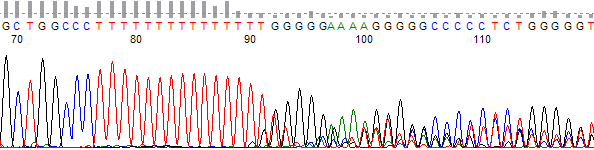
Figure 1. Polymerase slippage on a small mononucleotide T run.
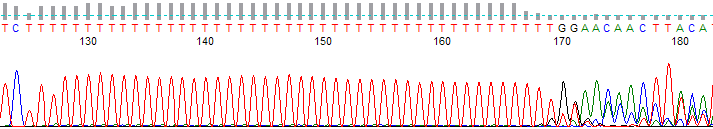
Figure 2. Polymerase slippage on a long mononucleotide T run.
Cause of DNA Sequence Slippage Problems
- Long runs of a mononucleotide base causes the DNA polymerase to “slip” on the template. The sequencing DNA polymerase skips over one or more bases make a product that is one or more bases shorter or longer. These cause of this slippage is due to the low processivity of the DNA polymerases combined with separation of the template and extension strand when the polymerase is not bound to the replication fork. When the strands come back together they can form a loop out effectively changing the lengths of some of the DNA products. When this happens it appears as mixed signal downstream of the mononucleotide run.
- Insertion and deletions in one allele. This can occur when two alleles are sequenced together (PCR templates) and one of the two mononucleotide runs has an insertion or deletion (indel) polymorphism.
Solving DNA Sequence Slippage Problems
- Sequence the DNA template from both directions. For example, if the template shows slippage after using the forward primer then sequence the template using a reverse primer. You may need to make a custom primers to do this if the template is large.
- Use a custom primer designed to hybridize just outside the mononucleotide or dinucleotide run region.
- Use the sequence-by-mutagenesis (SAM) approach to avoid have long mononucleotide runs in your templates.
Return to the main DNA sequencing troubleshooting page.