
PeakTrace™ is a novel DNA sequencing basecaller offering improved read lengths for DNA sequencing traces collected on the ABI 377, 310, 3700, 3730, 3730xl, 3100, 3130, 3200 (SeqStudio), 3500, and 3600 (SeqStudio Flex) DNA sequencers. The PeakTrace Basecalling software offers the following features.
- Extracts up to 50% more high quality bases per trace.
- Up to 3x increase in DNA sequencer throughput.
- Compatibility with all ABI DNA sequencers and downstream applications.
- Improved detection of polymorphic bases.
- Better trace appearance.
- Improved trace pass rate with fewer traces falling under 100 bases.
- Lower costs and increased sequencing instrument throughput.
- Available as both a standalone version (PeakTrace:Box) or an easy to use server version (Auto PeakTrace RP).
PeakTrace Basecalling Software
Automated DNA sequencers, like the ABI 3500 series, can collect sequencing reads that easily exceed 1400 nucleotides. Unfortunately, while the peak data is present in the trace file at the latter regions (post base 850), the peaks are broad and overlapping. The post base 850 region is associated with low quality scores and high basecall error rates making the post 850 base data of little use in most cases.
The PeakTrace Basecalling software uses a proprietary series of algorithms to extract additional high quality sequence data from the poorly resolved regions of each trace. The PeakTrace Basecaller extracts and reprocesses the raw data contained within each .ab1 trace file. This data reprocessing enables the PeakTrace Basecaller to make far better base and quality calls on the trace, as well as improving the appearance of the peaks.
An example of what the PeakTrace Basecaller can do is shown below (Figure 1).
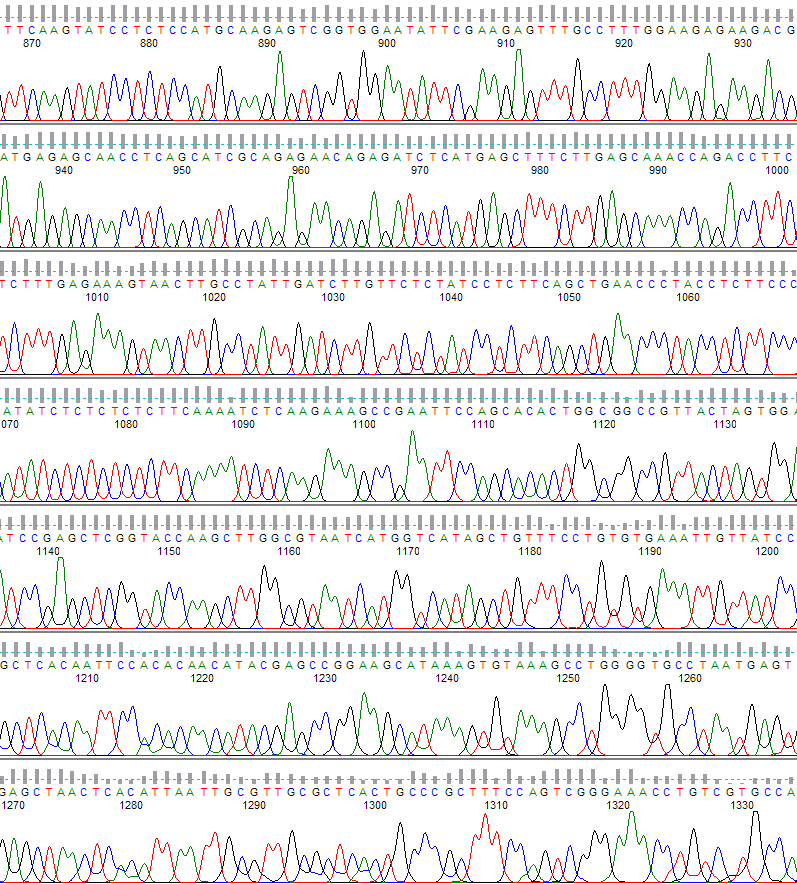
Figure 1. PeakTrace basecalling. The region shown is from base 870 to 1330.
Figure 2 is the same .ab1 file, but basecalled with the KB™ Basecaller.
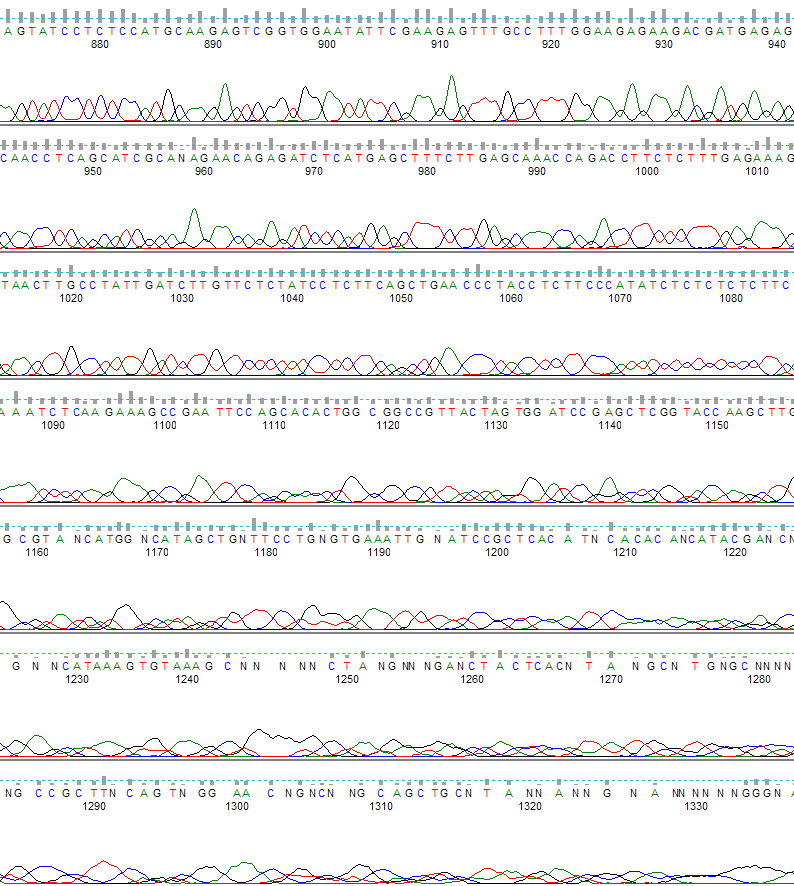
Figure 2. KB basecalling. The region shown is from base 870 to 1330.
PeakTrace basecalling can be performed any time after the trace data has been collected. This allows it to be used with previously obtained data sets such as partial genome sequencing projects data. By applying PeakTrace to such trace data and reassembling the sequences, the final genome assembly can be significantly improved without the need to collect more sequence data. Additionally, as the output of the PeakTrace basecaller is a standard .ab1 trace file, it means that PeakTrace can be used with standard downstream software applications such as Staden or Phrap.
Features of the PeakTrace Basecalling Software
Easy to use
PeakTrace is available in two major versions — an server version (Auto PeakTrace RP) and a fully offline version (PeakTrace:Box).
The Auto PeakTrace RP Basecalling software is very simple to use. After opening an Auto PeakTrace RP account and purchasing some processing units, users download the software and install it on their own computer. Users select the input folder containing the trace files and can set the basecalling settings via the options window. The traces are automatically uploaded to the PeakTrace server for processing. Once complete, the PeakTrace basecalled traces are downloaded by the RP software to the selected output folder on the users computer. Auto PeakTrace RP is available for Windows (XP, Vista, Win7, Win8, Win10).
The PeakTrace:Box Basecalling system is equally easy to use. The system comes with a USB key that controls licensing and allows the system to be run offline. The PeakTrace:Box Basecalling system comes as either a graphical (Auto PeakTrace) or command line version (peaktrace) to allow the automatic processing of the traces and/or integration into existing LIMS. In addition, the Auto PeakTrace software provides log reports making it straightforward to use in a core lab environment where users are charged on a per trace basis. The PeakTrace:Box system is available for Windows (XP, Vista, Win7, Win8, Win10) and x84_64 Linux.
The PeakTrace Basecaller is compatible with trace files collected from all ABI DNA sequencing instruments including the ABI 3700, ABI 310, ABI 377, ABI 3100, ABI 3130, ABI 3200, ABI 3600, ABI 3730, and ABI 3500 sequencerd. PeakTrace is also compatible with normal and rapid run traces collected at high run voltages (>12 kV).
The PeakTrace basecaller can be trialed on your own traces using the free PeakTrace Online basecalling service. This provides the same basecalling quality as the paid Auto PeakTrace versions, however, it is limited to processing 250 traces per day. If you have a large number of traces to process, then we recommend that you purchase a Auto PeakTrace RP account.
Increases trace read length and reduces basecalling errors
The PeakTrace Basecaller provides the user with significantly longer and higher quality reads (Figure 1). These improved read lengths mean that fewer sequencing reactions need to be performed to complete a sequencing project, thus saving both time and money.
The improved DNA sequencing read lengths provided by the PeakTrace basecaller increase sequencing throughput and significantly lower the overall per base cost. In addition, these longer read lengths enable sequencing projects (e.g. whole-genome, cDNA clones, etc) to be completed at lower sequence coverage levels, thus offering further cost- and time-savings.
For further information on the PeakTrace Basecaller including how to purchase or trial, please email us at {This email is obscured. Your must have javascript enabled to see it} or download the PeakTrace Whitepaper.