
The QualTrace II™ DNA sequencing QC analysis program enables users to rapidly detect sequencing problems that limit sequence read length. The QualTrace software offers the following features:
- Real-time analysis of trace files and detection of nine different sequencing problem types.
- Compatibility with trace files produced by both the ABI base callers on the ABI and MegaBACE DNA sequencers.
- Easy integration into existing sequencing data pipeline and bioinformatics infrastructure.
- An ideal means for troubleshooting DNA sequencing problems and optimizing production protocols.
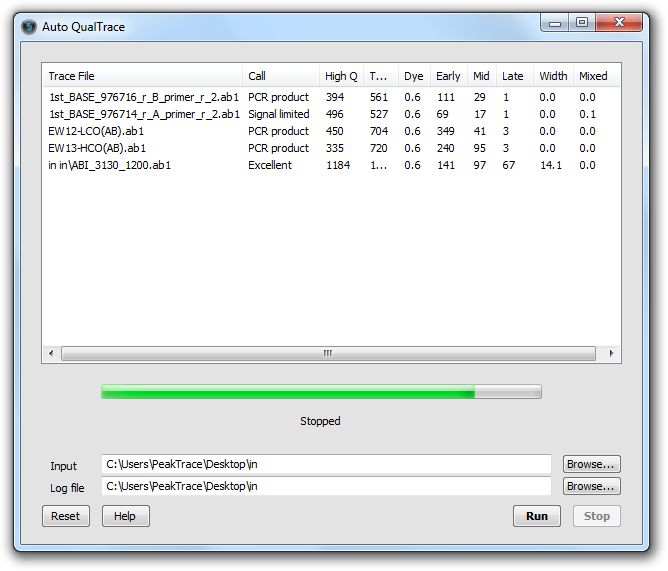
QualTrace II DNA Sequencing QC Program
The QualTrace II DNA sequencing software examines the raw dideoxy sequencing data contained within each ABI trace file to automatically identify nine problems with either the preparation of the DNA sequencing reactions, or with the DNA sequencer itself. The sequencing issues that can be identified by the QualTrace software include:
- Sequence traces that contain no signal data due to either a failed reaction or a blockage of the DNA sequencer capillary.
- Mixed trace signal resulting from multiple DNA templates in the sequencing reaction.
- Noisy or very low signal data traces that provide only short reads.
- Very weak signal strength at the end of the DNA sequencing trace resulting in short reads.
- Early mixed trace signal due to template contamination by PCR products.
- Delays in the starts of the DNA sequencing trace signal that indicate significant overloading of sequencer capillary with template DNA and/or other contaminants.
- Problems with the DNA sequencer’s spectral calibration resulting in major channel cross-talk signal.
- The presence of significant leftover BigDye sequencing mix in the loaded DNA sequencing reactions (dye blobs).
- Traces which show a rapid decline in peak signal.
- Indels (traces with insertions and deletions).
- Total number of high quality bases.
- Peak signal start.
- Instrument diagnostics such as machine name, run module, capillary length, run date, tray name, tray well, polymer lot, sample, sample id and user.
- The point in the traces where the trace signal reaches the noise threshold.
- The level of peak blur (poor peak resolution).
The QualTrace sequencing software can process either single .ab1 DNA trace files or folders of trace files. The software generates a log file report containing the QualTrace category type and code for each trace. In addition, the signal-to-noise ratio for the early, mid, and late regions of each trace are also recorded in the log file enabling the trace signal decay to be tracked. This ratio provides important information on ideal template concentration and BigDye mix, allowing effective optimization of the DNA sequencing process.
Use of QualTrace in DNA Sequencing Facilities
An example of the performance of QualTrace is illustrated in Table 1. A set of 11,745 ABI 3730xl traces of known sequence were collected using the ABI3XX base caller on a 36 cm array. The DNA traces were analyzed by QualTrace and sorted into their predicted categories. Each trace was base called using PHRED and the resulting sequences aligned to the known consensus sequence using CROSS_MATCH. The average number of aligned Q20+ phred bases for each of the nine QualTrace categories was calculated.
| QT category | Traces | Total Q20+ | Aligned Q20+ |
| Excellent trace | 7883 | 678 | 647 |
| Good trace | 773 | 627 | 606 |
| Spectral calibration needed | 87 | 664 | 609 |
| End weak traces | 65 | 566 | 479 |
| Mixed trace signal | 1453 | 342 | 322 |
| Early mixed trace signal | 521 | 531 | 508 |
| Noisy trace signal | 147 | 220 | 175 |
| Delayed start of trace signal | 1 | 27 | 0 |
| No signal or failed trace | 819 | 32 | 2 |
| Total | 11745 | 576 | 546 |
This analysis provided an accurate measurement of the predictive power of QualTrace II in identifying traces with production problems that limit alignable read length. The QualTrace DNA sequencing software was able to identify a significant problem in this data set of mixed signal resulting from multiple DNA templates (~17%). This problem was responsible for a major loss of DNA sequence read length with the average mixed trace providing only 322 phred Q20+ bases.
A second major issue identified by the QualTrace II DNA sequencing software was the large number (~7%) of failed traces. Continuous monitoring of this QualTrace failure type by capillary number allowed the identification of blocked capillaries that needed to be changed.
Auto QualTrace
A new graphical version of QualTrace II (Auto QualTrace) has been released, designed for small to medium sized DNA sequencing facilities (i.e. facilities with 1 – 10 ABI DNA sequencers). In addition to providing an easy to use interface, the Auto QualTrace offers a number of extra features aimed at providing low volume facilities access to the power of QualTrace QC analysis. For more information on Auto QualTrace please visit the Auto QualTrace overview page.
For more information about the QualTrace II or the Auto QualTrace II software please read the QualTrace DNA sequencing users manual, or email our support team at {This email is obscured. Your must have javascript enabled to see it} with any technical support questions you might have.
QualTrace II Ordering and Pricing Details
QualTrace II has been superseded by QualTrace III and is no longer sold or supported.