PeakTrace DNA Sequencing Basecaller
The PeakTrace™ Basecaller is a novel software system designed to improve the read length of sequencing traces from all ABI sequencing instruments. PeakTrace is compatible with traces generated by the ABI 377, ABI 310, ABI 3700, ABI 3100, ABI 3130, ABI 3500, ABI 3730, ABI 3730xl and MegaBACE sequencers. The PeakTrace basecaller is able to offer read length improvements of up to 100%.
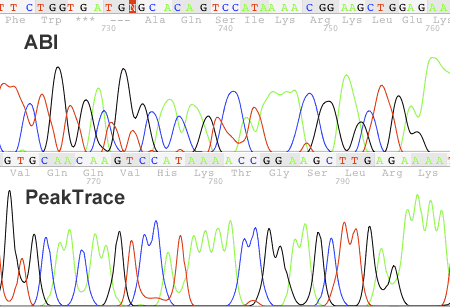
Figure 1. Example of PeakTrace Basecalling on a ABI 377 trace. The original ABI basecall is shown on top.
To try PeakTrace on your own traces and see what it can offer please visit our free online PeakTrace basecaller, the PeakTrace Basecaller, or PeakTrace FAQ page.
LongTrace DNA Sequencing Improvement System
The LongTrace DNA sequencing system is a new product for improving ABI 3730, 3730xl, 3100 and 3130 DNA sequencing traces. LongTrace works by reprocessing the raw sequencing data within the ABI trace file. This reprocessing allows the ABI KB basecaller to make much better base calls on the trace. This results in significant improvements in sequence read length by up to 30% or more for 3730 traces.
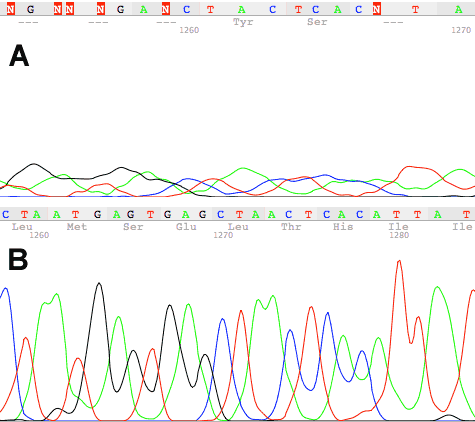
Figure 1. LongTrace reprocessing of a 50cm ABI 3130 DNA sequencing trace. A. The trace BEFORE LongTrace reprocessing (ie. KB base called). B. The same trace AFTER LongTrace reprocessing (ie. LongTrace processing then KB base calling). Note that the region shown from base 1260 to base 1285.
LongTrace has been superseded by PeakTrace and is no longer offered to new customers
dLUTE SEQ DNA Sequencing BigDye Dilution Reagent
The dLUTE family of reagent permits nanolitre scale molecular biological reactions to be performed using microlitre scale liquid handling equipment. The first dLUTE product to be released, dLUTE SEQ™, allows DNA sequencing reactions to be performed in volumes as small as 0.1µl using as little as 0.04µl (40nl) of the very expensive BigDye mix chemistry. This usage rate is equivalent to a 1 in 320 dilution sequencing reaction.

Figure 2. dLUTE SEQ before and after thermocycling. Left tube: The initial dLUTE state. In this state the aqueous phase (pink color) is dispersed throughout the inert phase. Right tube: The reaction dLUTE state. In this state the aqueous sequencing reaction phase (red) has separated from the inert phase (clear) which protects the reaction from evaporation.
The dLUTE SEQ sequencing reagent is compatible with all current robotic machinery making it ideal for DNA sequencing facilities wishing to reduce costs while retaining their current liquid handling equipment. The dLUTE reagent can be used on all modern DNA sequencers including the ABI 3700, ABI 3100, ABI 3130, ABI 3700, ABI 3730xl and the MegaBACE 500, MegaBACE 1000, MegaBACE 1500, & MegaBACE 4000.
The dLUTE SEQ reagent can also be used in PCR based applications as it allows the effective reaction volume to be reduced to as little as 0.1µl allowing great savings to also be made on expensive PCR reagents.
The formulation of dLUTE SEQ has been recently improved to make it easier to mix and remove the smell of the original version.
For further details and pricing information on the dLUTE SEQ reagent please visit the dLUTE SEQ DNA sequencing reagent page or download the dLUTE SEQ sequencing reagent overview pdf document.
We are currently developing a new formulation of dLUTE SEQ and until it is released we will no longer sell the older version.
dLUTE SEQ CF
We have developed a version of the dLUTE SEQ reagent (dLUTE SEQ CF) optimized for use by individual users and core facilities. This formulation provides small scale users a simpler reaction protocol as well as handling a wider range of template qualities and concentrations. For more information please visit the dLUTE SEQ CF overview page or our dLUTE SEQ users forum.
We are currently developing a new formulation of dLUTE SEQ CF and until it is released we will no longer sell the older version.
QualTrace DNA Sequence Trace Analysis Software
The QualTrace sequence analysis software provides real time detection of dideoxy-based automated sequencing production failures that can led to read length short falls. The QualTrace software’s analysis is independent of the base caller and can be used with the ABI 3730 and 3730xl DNA sequencers. In addition to providing valuable feedback on quality control issues, QualTrace is also ideal for troubleshooting the common problems that occur in large-scale sequencing projects.
The QualTrace DNA sequence analysis software works by analyzing the raw trace data contained within each ABI3730 DNA trace files. This enables the software to identify nine major sequencing problem types including:
- DNA traces that contain no data due to either failed sequencing reactions or blocked capillaries.
- The presence of mixed signal early in the trace signal indicating template contamination by PCR products or single primer PCR occurring in the sequencing reaction itself.
- Multiple sequencing peaks resulting from the presence of on or more DNA templates (eg. two plasmids from a double pick).
- Low signal at the end of the DNA trace that is at the instrument noise level.
- Delayed starts of the DNA trace signal indicating sample contamination issues with protein, excess DNA template or salt.
- The level of free BigDye mix reagent (dye blobs) present.
- Traces with very noisy signal data indicative of poor sequencing reactions and/or poor sample loading.
- Traces that show a rapid decline in signal strength (“ski slope” traces) indicative of excess template or primer imbalance.
- Problems with the ABI 3730 chemistry spectral calibration file.
In addition to being able to detect sequencing problems independent of the base caller used, the QualTrace™ analysis software also offers DNA sequencing core facilities a reproducible and cost efficient means of providing independent sequencing troubleshooting advice to their customers as to the cause of any sequencing problems.
The QualTrace program is available in versions designed for Windows XP, Vista, Win7 and Win8 and x86 Linux. It is designed to be integrated into current genome sequencing data pipeline and bioinformatics software systems. This makes it very easy to integrate QualTrace analysis into the production sequencing processes. A new graphical version of QualTrace (Auto QualTrace) has been released designed for small to medium scale sequencing facilities.
To allow users to see what the software offers we have created a free web version of QualTrace which users to see what the system can do on their own traces. To try the online version please visit the QualTrace online page.
For more information and prices please visit the QualTrace DNA sequencing software page, the QualTrace DNA sequencing software users manual, or download the QualTrace DNA sequencing software overview pdf document.
CounterTrace II DNA Sequencing Software & Reagent
CounterTrace II™ is an update of the CounterTrace DNA Sequencing Reagent & Software system designed for use with the ABI KB base caller for traces collected on the ABI 3730 and ABI 3730xl sequencers.
The CounterTrace II DNA sequencing software utilizes a labeled nucleic acid fragment ladder to enable improved signal processing of each sequence trace’s raw data. The CounterTrace standard is composed of DNA fragments of known size covering the base region from 520 to 1220 that are labeled with a proprietary fluorophore. This dye enables the standard peak locations to be detected and sized in the presence of the four color dideoxy sequencing peaks. The CounterTrace II DNA sequencing software uses the location of the CounterTrace standard peaks in the processing of the DNA trace data. The resulting improved automated DNA sequencing traces can be base and quality called using Applied Biosystems KB base caller.
The CounterTrace II DNA sequencing system offers increases in alignable KB Q20+ read lengths of up to 30% for individual sequence traces, with an average of 15 – 20% in high throughput sequencing centres. The CounterTrace II DNA sequencing software can also be used successfully in smaller core facilities and even by individual investigators.
For more details and pricing information please visit the CounterTrace II DNA sequencing software & reagent page or download the CounterTrace II overview.
CounterTrace II has been superseded by PeakTrace.
CounterTrace DNA Sequencing Software & Reagent
CounterTrace DNA sequencing system combines a propriety DNA standard with trace reprocessing software and can provides increases in trace read lengths of up to 65%, with average increases in the range of 25%. The CounterTrace sequencing kit is compatible with both the ABI 3700 and 3730 DNA sequencers and has been designed to be used with the phred base caller. CounterTrace requires only minimal changes of current DNA sequencing protocols to utilize.
For more information about CounterTrace please visit the CounterTrace DNA sequencing software & reagent page, or download the CounterTrace overview.
UniSeq Genome Walking Oligonucleotide Primer Library
The UniSeq™ genome DNA sequencing technology allows the user to rapidly sequence any cloned DNA region by the primer walking method without the need for custom oligonucleotide synthesis. The UniSeq technology is based on the UniPrime™ technology developed by Nucleics. UniPrime utilizes a unique design and combination of short oligonucleotides to produce individual, template-specific, sequencing primers. These primers are used directly in the Sanger DNA sequencing reaction without the need for additional enzymatic steps.
The associated software (UniFinish™) can rapidly parse genome assembly files (e.g. PHRAP ace files) and identify the DNA templates and oligonucleotide primer pairs required to close contig gaps and poor quality regions by the primer walking method. This software allows easy integration into existing whole genome sequencing shotgun operations. The UniSeq technology is robust and provides significant time and cost savings over current genome sequencing protocols.
For further information on the UniSeq™ system please visit the UniSeq page download the UniSeq genome sequencing overview or visit the UniSeq DNA sequencing FAQ page.
ASIN Genomic DNA Sequencing System
ASIN is a novel DNA amplification and sanger sequencing system developed by Nucleics. This technology enables cost efficient DNA sequencing of any PCR amplifiable DNA or RNA template. The ASIN approach utilizes oligonucleotide primers selected from a universal pre-synthesised kit of only 400 oligonucleotides. An advantage of the ASIN technology is its compatibility with all current sequencing chemistries, including automated Sanger DNA sequencing, manual DNA sequencing and even Maxam and Gilbert chemical sequencing. The nature of ASIN makes it suitability for automation.
The ASIN genome sequencing technology was sold to Takara Biomedicals, a division of Takara Shuzo (Japan) in February 2000.