Identification of chimera and rearranged DNA sequences
- The trace is high quality up to a certain point, then becomes mixed. Sometimes the mixed signals appear to be related to each other in that the peaks present are ‘echoed’ later in the trace.
- The trace sequence is excellent, but the basecall data only aligns to the expected data up to a point and then shows no further homology.
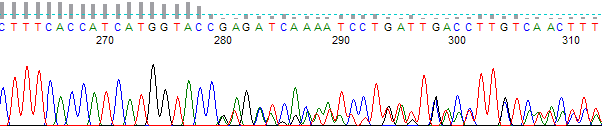
Figure 1. Example of a chimeric trace
Causes of chimera and rearranged sequences
- Cloning artifacts. Chimeras can result from cloning two or more unrelated DNA fragments at once in the same vector. This is more likely to occur with some cloning techniques such as restriction enzyme digested inserts.
- ‘Unstable’ sequences such as sequence repeats or long mononucleotide runs. This is a common problem when cloning cDNA, as the long poly A tail is unstable in many vectors or E. coli strains.
- ‘Toxic’ sequences. If the gene cloned is expressed in E. coli and is toxic to the cell, then you will select for clones that have deleted or rearranged the toxic region. This problem tends to occur more commonly with high copy number vectors and templates with low G+C ratios.
- Strong secondary structure. Deletion can occur in regions of strong secondary structure. This is commonly seen when PCR products are cloned, as the PCR process preferentially amplifies for deletion artefacts that remove the secondary structure.
- Double picks. This is where two colonies are isolated that have grown close to each other such that you end up with two templates.
Solutions to chimera and rearranged sequences
- Isolate and sequence another clone. This can solve the problem if the particular clone you have used has undergone a rare mutation event. Sometimes going back to the original plate and reselecting the same colony can solve the problem, as the mutation may have occurred in later liquid cultures.
- Use a low copy number vector and/or grow the cells at 30˚C. This can help maintain the stability of unstable or toxic sequences by reducing the selective pressure for mutation or rearrangement. Growing at lower temperatures helps stop the cells overgrowing.
- Don’t overgrow your cultures. The longer the cells are grown, the higher the selection pressure for a deletion or rearrangement mutant to arise. This effect is particularly strong in liquid culture.
- Try cloning using a different E. coli strain. This can sometimes work, especially if you originally used a non-cloning strain such as BL21.
- Try adding some glucose (50 − 100 mM) to the growth media to inhibit expression of the lacZ promoter. This can help if the instability is caused by leaky expression of a gene under the lacZ promoter.
Return to the main DNA sequencing troubleshooting page.
