Identification of sequencing trace signal hard stops
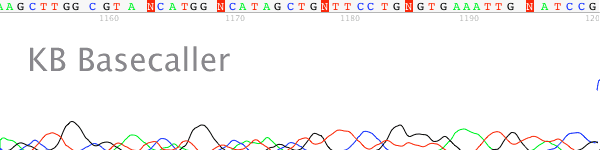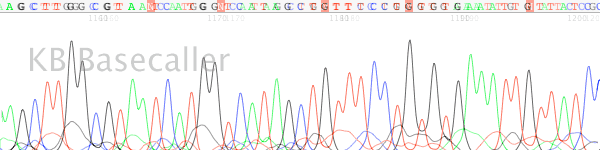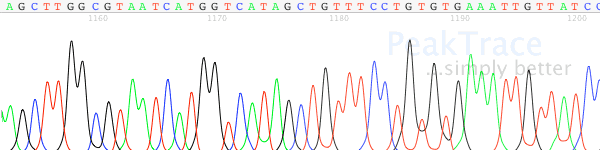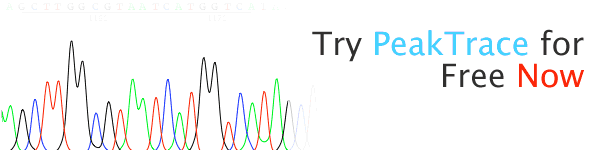
- Good quality trace signal suddenly stops, or rapidly declines, in regions of the traces that should be well resolved (Figure 1).
- The trace peak signal falls to under 150 units (on ABI 3730 instruments) in the raw channel after the stop region (Figure 2).
- The average quality scores after the “hard stop” region are generally low.
- Template has a high percentage of G (Figures 1, 2, 3 and 4) and/or C nucleotides (Figures 5 and 6), especially in the region where the stop occurred.
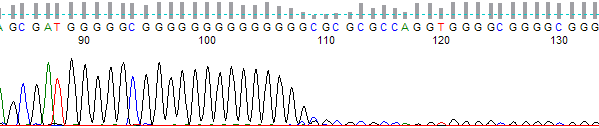
Figure 1. Processed output showing a sudden stop in the trace after a long G run
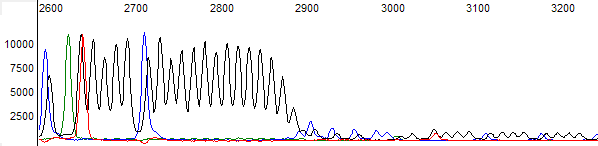
Figure 2. The raw channel data of the trace shown in figure 1. Note the extreme signal drop off after the G run region
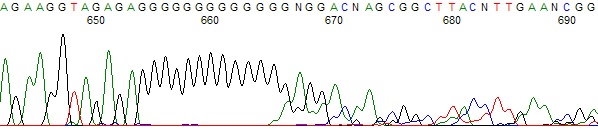
Figure 3. Signal drop off after long G run
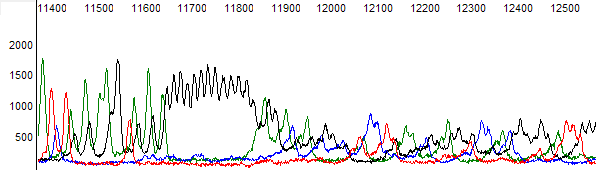
Figure 4. The raw channel data of the Figure 3 processed trace. After the G run the trace loses resolution and noise levels increase.
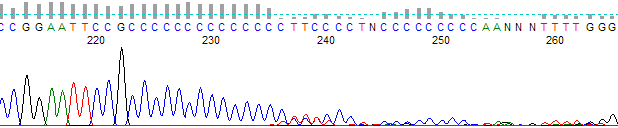
Figure 5. Hard stop after a C run
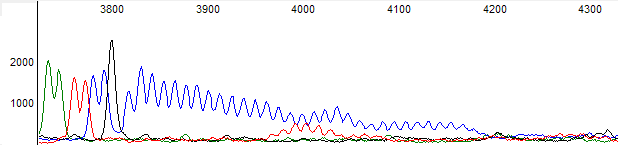
Figure 6. Raw channel data for figure 5 trace.
Causes of sequencing trace signal sudden stops
- Regions of strong secondary structure in the DNA template. These regions can fold back on themselves and form hairpin and knot-like structures that the DNA sequencing polymerase can not pass through.
- Long runs of G or C nucleotides in the template (Figures 1, 2, 3, and 4). These can not only form hairpins with strong secondary structure, but can also form single stranded conformational structures that the DNA sequencing polymerase has difficulty passing through. Long template regions of guanidine (G runs) are particularly problematic because of the substitution of dITP for dGTP in the BigDye sequencing mix. The BigDye sequencing DNA polymerase does not efficiently extend runs of multiple inosine residues, causing the polymerase to stop in long G run regions.
Solutions for overcoming DNA sequencing hard stops
- Use a 1:4 ratio mix of BigDye™ to dGTP Sequencing premix. This mix will help sequence through high G+C templates.
- Add a small amount of dGTP nucleotide (~40µM) to the BigDye™ mix. This helps the sequencing polymerase pass through long G runs without causing major compression problems.
- PCR amplify the template using 7-deaza-GTP or dITP to reduce the strength of the secondary structure. This allows the sequencing polymerase to be able to pass through the hairpin region causing the hard stop.
- Use the sequence-by-mutagenesis (SAM) approach to avoid having long mononucleotide runs in your templates. This SAM approach is very clever and provides a lot of advantages for all sorts of problematic templates.
For information on the real time detection of DNA sequencing problems please visit the QualTrace DNA sequencing analysis software page.