Identification of PCR/Primer Dimer Problems in Traces
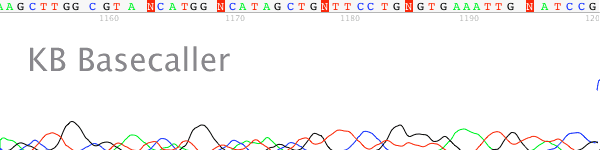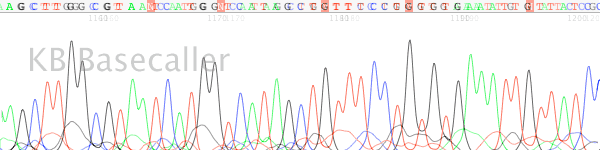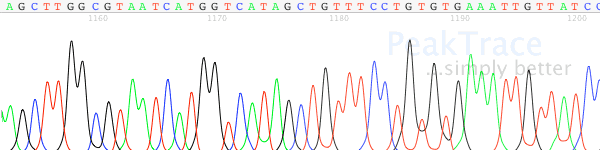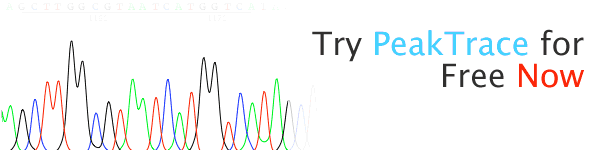
- The trace signal is mixed in the early regions (normally before base 200), yet the later regions are not mixed. There is typically a sharp transition between the two regions (Figure 1).
- The signal drops from very high early in the trace (usually in the first 200 bases) to much lower levels. Examination of the raw signal shows a high signal “block” (Figure 2).
- The total quality scores counts are generally low to moderate depending on the size of the region affected.
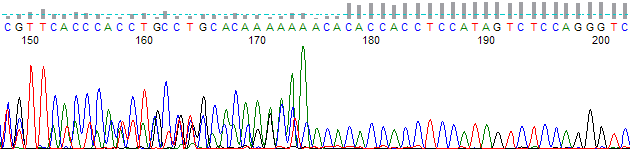
Figure 1. Transition from mixed to non-mixed sequence at base =175.
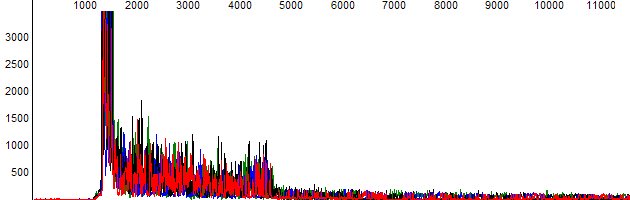
Figure 2. High signal block at the start of the trace as seen in the raw data channel.
Causes of PCR/Primer Dimers in Sequencing Reactions
- Contamination of the template, primer stock or other sequencing reagents with primer dimers.
- Too low an annealing temperature during the PCR.
- Two primer binding sites present in the template.
- Direct sequencing of PCR products where there is more than one band.
- Sequencing TempliPhi™ templates. For reasons unknown TempliPhi templates are much more prone to primer dimer problems.
Solving Primer Dimer Problems
- Change all the sequencing reagents and water for a fresh batch. Also clean the pipettors as they can also be the cause of the primer dimer contamination.
- Raise the annealing temperature and/or use a shorter annealing time.
- Use different primers for sequencing that is internal to the PCR primer sites (i.e. nested). It is not a good idea to use the same primers to sequence with as you used to amplify the PCR product.
- Check the sequence of the vector to ensure that there are not two primer binding sites present. Be especially careful of partial matches – an 8-9 base pair match can cause problems if at the 3′ end of the primer.
For more information on detecting DNA sequencing trace problems please visit the QualTrace III DNA sequencing analysis software page.
Return to the main DNA sequencing troubleshooting page.