QualTrace III is an integral part of PeakTrace 6 and can be used as either part of the standard PeakTrace basecalling, or on its own via selecting qualtrace as the basecaller within Auto PeakTrace 6.
QualTrace III Options Window
The QualTrace III options allow the default QualTrace III processing options to be adjusted as well as allowing the detail level of the Trace Reports to be set.
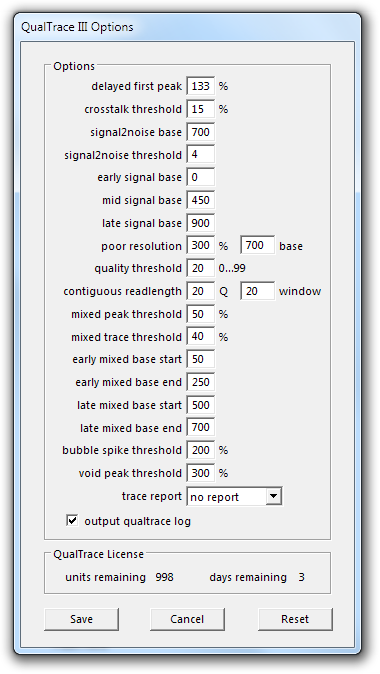
QualTrace and Trace Report Optional Parameters
The setting on this menu allow the user to override the default settings for QualTrace. For most of these options it is strongly advised to not change these settings (other than the trace report setting) as the maximum utility is obtained with QualTrace when using consistent values allow historical comparisons to be made. This can enable drift in protocols or regent decay to be detected early. In addition, using inappropriate values may result in invalid analysis results.
delayed first peak
The percentage delay of the actual first peak from the expected for the trace to be classified as Late Peak Start. Default is 133%.
crosstalk threshold
The percentage of spectral crosstalk (bleed of signal from one channel into other channels) due to mis-calibration of DNA sequencing instrument for a trace to be classified as Spectral Calibration. Default is 15%.
signal2noise base
The approximate base from which signal to noise measurements are made to determine if a trace falls into the Excellent, Good, Below average, Poor, or Failed categories. Default is 700.
signal2noise threshold
The signal to noise ratio used to determine if a trace falls into the Excellent, Good, Below average, Poor, or Failed categories. Default is 4.
early signal base
The base (to the mid signal base) from which the early signal to noise ratio (EARLY SIGNAL) is determined. Default is 0.
mid signal base
The base (to the late signal base) from which the mid signal to noise ratio (MID SIGNAL) is determined. Default is 450.
late signal base
The base (to the end of the basecall) from which the late signal to noise ratio (LATE SIGNAL) is determined. Default is 900.
poor resolution
The measured peak width over the expected width at the set poor resolution base to classify a trace as Poor Resolution. Default is 300% and base 700.
quality threshold
The minimum quality score used to calculate GOOD BASE. Default is 20.
contiguous readlength
The minimum quality score threshold and window size used to calculate CRL. Default is 20 and 20.
mixed peak threshold
The percentage height of the second peak beneath the primary peak required to classify a peak as mixed signal. Default is 50%.
mixed trace threshold
The percentage of mix peaks is the mixed base window to classify a trace as Early Mixed or Late Mixed. Default is 40%.
early mixed base start
The start base of the early mixed base region. Default is 50.
early mix base end
The end base of the early mixed base region. Default 250.
late mixed base start
The start base of the late mixed base region. Default is 500.
late mixed base end
The end base of late mixed base region. Default is 700.
bubble spike threshold
The percentage height that a narrow peak with strong signal in all 4 channels must be above the average peak height to detect the presence of air bubbles (BUBBLE). Default is 200%.
void peak threshold
The percentage height that a broad peak with strong signal in all 4 channels must be above the average peak height for the void peak base (VOID PEAK) to be calculated. Default is 300%.
trace report
This option controls the detail of trace report. The four options are no report, brief, standard and full. Default is no report.
output qualtrace log
This option controls if a qualtrace log file is generated. The tab delimited logfile is saved to the output folder with the name “qualtrace.xls”. Default is checked (QualTrace log file output).
QualTrace III Log and Trace Reports
QualTrace III output information of 55 trace parameters covering a wide range of run conditions and trace quality. The categories reported are.
QT CODE
A numerical classification of the trace by QualTrace III. Most codes are shared with PeakTrace with the following additional codes.
- 40 Excellent trace
- 41 Good trace
- 42 Data collected limited
- 43 Below average trace
- 44 Poor trace
- 45 Late peak start
- 46 Spectral calibration
- 47 PCR product
- 48 Early mixed peaks
- 49 Late mixed peaks
- 50 Poor resolution
QT CLASS
Excellent Trace
The raw channel signal falls after the signal2noise threshold (default 4) after the signal2noise base (default 700) plus 250. Peak signal strength is not the limiting factor for trace read length. Production protocols should be optimized to achieve this results.
Alignable Q20+ bases are typically high to very high.
Good Trace
The raw channel signal falls after the signal2noise threshold (default 4) after the signal2noise base (default 700), but before signal2noise base plus 250. Peak signal strength is the limiting factor for trace read length.
Alignable Q20+ bases are typically high.
Data collected limited
The data collection stops before the raw channel signal falls below the signal2noise threshold (default 4). The read length is limited because the run was stopped too soon. This problem can be solved by increasing the run time.
Below Average Trace
The raw channel signal falls below the signal2noise threshold (default 4) before the signal2noise base (default 700), but after signal2noise base minus 400. Peak signal strength is a major limiting factor for trace read length.
- Incorrect template concentration.
- Excessive sequencing chemistry dilution.
- DNA sequencing instrument failure.
- Blocked capillaries.
- Contamination with salts, detergents, proteins, RNA, or excessive DNA.
Alignable Q20+ bases are typically low to moderate.
Poor Trace
The raw channel signal falls below the signal2noise threshold (default 4) 400 bases before the signal2noise base (default 700). This result can be caused by:
- Incorrect template concentration.
- Excessive sequencing chemistry dilution.
- DNA sequencing instrument failure.
- Blocked capillaries.
- Contamination with salts, detergents, proteins, RNA, or excessive DNA.
Alignable Q20+ bases are low.
Failed Trace
Trace file contains no or very weak peak signal in the raw data channels (failed reaction). Any signal present has fallen below the signal2noise threshold (default 4) before base 100. This result can be caused by:
- No DNA template.
- Loss of sample during clean up.
- A blocked capillary.
- Heavy contamination of the template DNA with salt or other charged ions.
Alignable Q20+ bases are very low to zero.
PCR Product
Trace file appears to be derived from a PCR product that results in the peak signal ending. The read length is limited by the length of the PCR product.
Alignable Q20+ bases are variable as they depend on the length of the PCR product.
Early Mixed Trace
More than the mixed peak threshold (default 40%) of the primary peaks from early mixed base start (default 50) to early mixed base end (default 250) contain a secondary peak more than the mixed peak threshold (default 50%) of the height of the primary peak. The peaks in the later regions of the trace are often good with only the beginning of the trace mixed. This result can be caused by:
- Contamination of the template with a short PCR product from PCR primer mis-annealing.
- Two primer binding sites in the template
- Primer mis-annealing at secondary sites within the template
- Single primer PCR amplification in the sequencing reaction.
Alignable Q20+ bases are typically moderate to high.
Late Mixed Trace
More than the mixed peak threshold (default 40%) of the primary peaks from late mixed base start (default 500) to late mixed base end (default 700) contain a secondary peak more than the mixed peak threshold (default 50%) of the height of the primary peak. This result can be caused by:
- Two different DNA templates present in the sequencing reaction.
- Two primer binding sites in the template
- Primer mis-annealing at secondary sites within the template
- Single primer PCR amplification in the sequencing reaction.
Alignable Q20+ bases are low or moderate.
Late Peak Start
The first signal peak collection has been time delayed by 33% compared to the expected collection time for this trace run condition. This error can occur when the capillary is overloaded with protein, template DNA, or salt.
Alignable Q20+ bases are moderate to high.
Spectral Calibration
Indicates that the trace file contains significant fluorescent dye channel signal crosstalk (a minimum of 15% in at least one channel). Multiple occurrences of this result for an individual DNA sequencer or capillary suggest that a machine spectral calibration should be performed.
Alignable Q20+ bases are moderate to high depending on the severity of the signal cross talk.
Poor Resolution
The peak width at poor resolution base (default 700) is poor resolution percentage (default 300) broader than expected for traces collected under the current run condition. Excessive peak widths are indicative of poor peak resolution and can result from:
- Excessive template DNA.
- DNA sequencing instrument failure.
- Failed arrays that need to be cleaned or replaced.
- Contamination with salts, detergents, proteins, RNA, or non-template DNA.
Alignable Q20+ bases are moderate to low.
Unsupported Run Condition
QualTrace III does not currently support all possible DNA sequencer run conditions. This result can occur if using custom modified run conditions, or because of an unusually combinations of run voltage, polymer type, instrument, dye chemistry, and/or capillary lengths. If you obtain this result please contact Nucleics as we can add support for new run conditions quickly once we have example trace files. The following run conditions are currently supported:
ABI 3730, 3730xl, 3100 and 3130 Run Conditions
- POP7, BigDye3, 8.5kV, 50cm array traces
- POP7, BigDye1, 8.5kV, 50cm array traces
- POP7, BigDye3, 8.5kV, 36cm array traces
- POP7, BigDye1, 8.5kV, 36cm array traces
- POP7, BigDye3, 6.1kV (long run), 50cm array traces
- POP7, BigDye1, 6.1kV (long run), 50cm array traces
- POP7, BigDye3, >12kV (rapid run), 50cm array traces
- POP7, BigDye1, >12kV (rapid run), 50cm array traces
- POP7, BigDye3, >12kV (rapid run), 36cm array traces
- POP7, BigDye1, >12kV (rapid run), 36cm array traces
- POP7, ET Terminator, 8.5kV, 50cm array traces
- POP7, ET Terminator, 8.5kV, 36cm array traces
- POP7, BigDye3, 8.5kV, 80cm array traces
- POP7, BigDye1, 8.5kV, 80cm array traces
- POP6, BigDye3, 8.5kV, 50cm array traces
- POP6, BigDye1, 8.5kV, 50cm array traces
- POP6, BigDye3, 8.5kV, 36cm array traces
- POP6, BigDye1, 8.5kV, 36cm array traces
- POP6, ET Terminator, 8.5kV, 50cm array traces
- POP6, BigDye3, >12kV (rapid run), 50cm array traces
- POP6, BigDye1, >12kV (rapid run), 50cm array traces
- POP6, BigDye3, >12kV (rapid run), 36cm array traces
- POP6, BigDye1, >12kV (rapid run), 36cm array traces
- POP4, BigDye3, 8.5kV, 80cm array traces
- POP4, BigDye1, 8.5kV, 80cm array traces
- POP4, BigDye3, 8.5kV, 50cm array traces
- POP4, BigDye1, 8.5kV, 50cm array traces
- POP4, BigDye3, 8.5kV, 36cm array traces
- POP4, BigDye3, 8.5kV, 36cm array traces
- POP4, BigDye1, 8.5kV, 36cm array traces
- POP4, BigDye3, >12kV (rapid run), 36cm array traces
ABI 3500 Run Conditions
- POP7, BigDye3, 8.5kV, 50cm array traces
- POP7, BigDye1, 8.5kV, 50cm array traces
- POP7, BigDye3, >13kV (rapid run), 50cm array traces
- POP7, BigDye1, >13kV (rapid run), 50cm array traces
ABI 3700 Run Conditions
- POP5, BigDye3, 50cm array traces
- POP5, BigDye1, 50cm array traces
- POP6, BigDye3, 50cm array traces
- POP6, BigDye1, 50cm array traces
ABI 310 Run Conditions
- POP6, BigDye3, 50cm array traces
- POP6, BigDye1, 50cm array traces
- POP6, BigDye3, 36cm array traces
- POP6, BigDye1, 36cm array traces
- POP4, BigDye3, 36cm array traces
- POP6, BigDye3, 30cm array traces
- POP6, BigDye1, 30cm array traces
- POP4, BigDye3, 30cm array traces
- POP4, BigDye1, 30cm array traces
ABI 377 Run Conditions
- BigDye3 traces
- BigDye1 traces
- ET Terminator traces
MegaBACE Run Conditions
- BigDye3 traces
- ET Terminator traces
- ET Primer traces
MJ Research BioStation Run Conditions
- Dye Rhodamine traces
QT DATE
The time and date the QualTrace III analysis was performed.
FILE
Name of the trace file.
MACHINE
The name of the DNA sequencing instrument the trace was collected on as recorded in the .ab1 field MCHN.
MODULE
The name of the run module the trace was collected using as recorded in the .ab1 field MODF.
RUN
The run name on as recorded in the .ab1 field RunN.
RUN DATE
Date and time the trace file was collected.
BASECALLER
The basecaller and version number used to basecall the trace file as recorded in the .ab1 field SVER.
CAPILLARY
The array capillary number the trace was collected using as recorded in the .ab1 field LANE.
LENGTH
The length of the capillary array in cm as recorded in the .ab1 field LNTD.
POLYMER LOT
The polymer lot used as recorded in the .ab1 field SMLt.
TRAY NAME
The name of the tray the sample was loaded from as recorded in the .ab1 field CTNM.
TRAY WELL
The tray well the sample was loaded from as recorded in the .ab1 field TUBE.
SAMPLE ID
The sample id as recorded in the .ab1 field LIMS.
SAMPLE
The sample name as recorded in the .ab1 field SMPL.
COMMENT
The sample comment as recorded in the .ab1 field CMNT.
OWNER
The owner of the sample as recorded in the .ab1 field CTOw.
USER
The DNA sequencing instrument operator as recorded in the .ab1 field User.
SET TRAY TEMP
The buffer tray temperature as set in the ABI Data Collection software in degrees C.
SET DETECTOR TEMP
The detector cell temperature as set in the ABI Data Collection software in degrees C.
SET RUN TEMP
The run temperature as set in the ABI Data Collection software in degrees C.
SET RUN VOLT
The run voltage as set in the ABI Data Collection software in kV.
RATE
The scan rate as set in the ABI Data Collection software. Values indicate time between scans in seconds.
LASER
The laser power setting as recorded in the .ab1 field LsrP in microWatts.
RUN VOLT
Median voltage as observed during the run in kV.
RUN CURRENT
Median current as observed during the run in milliAmps.
RUN POWER
Median power as observed during the run in milliWatts.
RUN TEMP
Median temperature as observed during the run in degrees C.
INJECT VOLT
The injection voltage used in kV.
INJECT TIME
The injection time in seconds.
SPACING
The inter peak spacing as recorded in the .ab1 field SPAC.
RUN TIME
The time of data collection in seconds.
SCANS
The number of scans collected as recorded in the .ab1 field SCAN.
DETECTOR OVERLOAD
The number of data points in the raw channels that exceed 32000. Values about 0 indicate that too much DNA was loaded.
TRACE SCORE
The average quality score of the trace within the CRL. This score is analogous to sample score provided by the ABI Sequence Analysis Report Manager.
CRL
The Contiguous Range Length (CRL) is the longest uninterrupted stretch of bases with an average quality score higher than the contiguous readlength quality value (default 20) using the contiguous readlength sliding window (default 20). This score is analogous to LOR value provided by the ABI Sequence Analysis Report Manager.
NOISE
The average noise value across all four raw data channels.
SPECTRAL
The crosstalk signal in the channel showing the greatest level of signal crosstalk (i.e. cross signal in one channel due to strong signal in another channel). Values above 0.1 indicate that a spectral calibration should be performed on the instrument.
DYE SIGNAL
The ratio of the largest potential “dye blob” peak to the average sequencing peak signal. Values greater than 1 are indicative of the presence of dye blobs. Values above 4 indicate major dye blob peaks are present.
BUBBLE
The number of narrow peaks (spikes) with signal in all 4 raw channels where the spike is greater than the bubble spike threshold (default 200%) more than the median peak signal height. Bubble spikes result from small gas bubbles in the polymer moving past the detector.
VOID PEAK
The number of late peaks with the signal in all four channels is void peak threshold more than the median signal. Values above 0 indicate the trace collection time was excessive for the run voltage and polymer used.
PEAK START
The scan number of the first sequence peak as recorded in the .ab1 field B1Pt.
EARLY SIGNAL
The signal-to-noise ratio of the first region between the early signal base (default 0) and the mid signal base (default 450) of the raw data channels.
MID SIGNAL
The signal-to-noise ratio of the second region between the mid signal base (default 450) and the late signal base (default 900) of the raw data channels.
LATE SIGNAL
The signal-to-noise ratio of the third region between the late signal base (default 900) and the last data point of the raw data channels.
NOISE START
The approximate base number where the trace signal-to-noise ratio falls below the signal2noise threshold (default 4).
QCUT
The quality score threshold for a high quality base as set by the quality threshold value (default 20).
INDEL
Indicates if the trace contains an insertion or deletion. A value of 0 indicates no indel detected, any number above 0 indicates the approximate base position of the suspected first indel.
TOTAL BASES
The total number of basecalled nucleotides in the trace file.
GOOD BASES
The total number of basecalls with quality scores greater than or equal to the quality threshold.
EARLY MIX
The ratio of those primary peaks which have an underlying secondary peak of more than the mixed peak threshold (default 50%) of the height primary peak to the total number of primary peaks in the region from early mixed base start (default 50) to early mixed base end (default 250). Values above 0.1 are indicative of mixed signal in the early region of the trace.
LATE MIX
The ratio of those primary peaks which have an underlying secondary peak of more than the mixed peak threshold (default 50%) of the height primary peak to the total number of primary peaks in the region from late mixed base start (default 500) to late mixed base end (default 700). Values above 0.1 are indicative of mixed signal in the late region of the trace.
PEAK WIDTH
The width of the peak at the poor resolution base (default 700) at half the peak height. The larger this value the lower the peak resolution is for a given run condition. Large values relatively to the SPACING value indicate low quality traces with poor peaks resolution.
QualTrace III Errors
QualTrace III shares the same error codes with PeakTrace. The single error unique to QualTrace III is
qualtrace license expired
Indicates that the QualTrace III license has expired. Please contact Nucleics to obtain a license upgrade.
Trace Reports
QualTrace III can also generate text based Trace Report files. This files can be supplied to end users to explain to them why their trace is not the quality they are expecting and what they might do to correct the problem(s). The report files can be generated with increase levels of report detail depending on end user needs. The report files are named after the trace file with “_report.txt” added (e.g. if the trace file name is “trace123.ab1” then the trace report file will be named “trace123_report.txt”) and saved to the same folder as the output trace file.
The four Trace Report options and outputs are:
No Report
This option prevents the output of a trace report. This is the default value.
Brief
Outputs a short description of the most important QualTrace III analysis results. An example is shown below.
SUMMARY
Trace File: ABI_3130_1200_B.ab1
PeakTrace Class: Success
Total Bases: 1367
KB Good Bases: 1333
PT Good Bases: 1340
Improvement: 0.5%
Date: 2015/08/11 10:55:00
Trace Score: 50
Contiguous Read Length: 1344
Standard
Outputs a summary and detailed description of the important QualTrace III analysis results. An example is shown below.
SUMMARY
Trace File: ABI_3130_1200_B.ab1
PeakTrace Class: Success
Total Bases: 1367
KB Good Bases: 1333
PT Good Bases: 1340
Improvement: 0.5%
Date: 2015/08/11 10:57:38
DETAILS
QualTrace Class: Excellent trace
Average Peak Spacing: 14.41
Run Time: 9000 sec.
Total Scans: 17960
Detector Overload: 0
Trace Score: 50
Contiguous Read Length: 1344
Dye Signal: 0.94
Bubble Spike(s): 0
Void Peak(s): 0
First Peak at Scan: 1276
Early Signal to Noise: 80
Mid Signal to Noise: 64
Late Signal to Noise: 46
Signal at Noise Level at Base: 1406
InDel(s): 0
Early Mixed Peaks: 0.00
Late Mixed Peaks:0.00
Peak Width at Base 700: 13.29
Full
Outputs the summary and full details of the QualTrace III analysis in text form including the PeakTrace and QualTrace III settings used for the analysis. An example is shown below.
SUMMARY
Trace File: ABI_3130_1200_B.ab1
PeakTrace Class: Success
PeakTrace Code: 0
Total Bases: 1367
KB Good Bases: 1333
PT Good Bases: 1340
Improvement: 0.5%
Date: 2015/08/11 11:00:21
DETAILS
QualTrace Class: Excellent trace
QualTrace Code: 40
Sequencer: 3730-16112-027-16112-027
Run Module: XLRSeq50_POP7_1
Run: Run_3730-16112-027_2007-03-17_16-00_0297
Run Date: 2007/03/17 17:00:52
Original Basecaller: PT 5.51
Capillary Number: 23
Capillary Length: 50
Polymer Lot: 0701015
Tray: 343
Tray Well: B5
Sample ID: 58e5d004d3f311ebaabf000d56d7a0f8
Sample Name: 23997
Sample Comment: <ID:23997>
Sample Owner: Daniel_Tillett
Sequencer User: Daniel_Tillett
Run Temperature Set: 60 deg.C
Run Voltage Set: 6.10 kV
Scan Rate: 0.25 sec.
Measured Run Voltage: 6.0 kV
Measured Run Current: 143.0 mA
Measured Run Power: 25.0 mW
Measured Run Temperature: 60.0 deg.C
Injection Voltage: 15.0 kV
Injection Time: 15 sec.
Average Peak Spacing: 14.41
Run Time: 9000 sec.
Total Scans: 17960
Detector Overload: 0
Trace Score: 50
Contiguous Read Length: 1344
Average Detector Noise: 5.21
Spectral Calibration: 0.08
Dye Signal: 0.94
Bubble Spike(s): 0
Void Peak(s): 0
First Peak at Scan: 1276
Early Signal to Noise: 80
Mid Signal to Noise: 64
Late Signal to Noise: 46
Input Total Bases: 1406
Signal at Noise Level at Base: 1406
Quality Score Threshold: 20
InDel(s): 0
Early Mixed Peaks: 0.00
Late Mixed Peaks: 0.00
Peak Width at Base 700: 13.29
PEAKTRACE SETTINGS
Trace File Output: ab1
Seq File Output: abi
.phd.1 file output: no phd
Good Base Improvement: -20
Good Quality Threshold: 20
N Base Threshold: 5
Mixed Peak Threshold: 0
Signal Start Peak: auto
Basecaller: peaktrace
Fuse Trace at Base: 20
Q Average Trim: 12 20
Force Processing: no
True Profile: no
Skip Short Traces: no
Trim Improved Traces: yes
Stealth Mode: no
Noise Raw Data: no
No Peak Resolution: no
Clean Baseline: no
Extra Normalization: no
QUALTRACE SETTINGS
Delayed First Peak: 133%
Crosstalk Threshold: 15%
Signal to Noise Base: 700
Signal to Noise Threshold: 4
Early Signal Base: 80
Mid Signal Base: 64
Late Signal Base: 46
Poor Resolution Threshold: 300%
Poor Resolution Base: 700
Quality Threshold: 20
Contiguous Readlength: 20 20
Mixed Peak Threshold: 50%
Mixed Trace Threshold: 40%
Early Mixed Base Start: 50
Early Mixed Base End: 250
Late Mixed Base Start: 500
Late Mixed Base End: 700
Bubble Spike Threshold: 200%
Void Peak Threshold: 300%