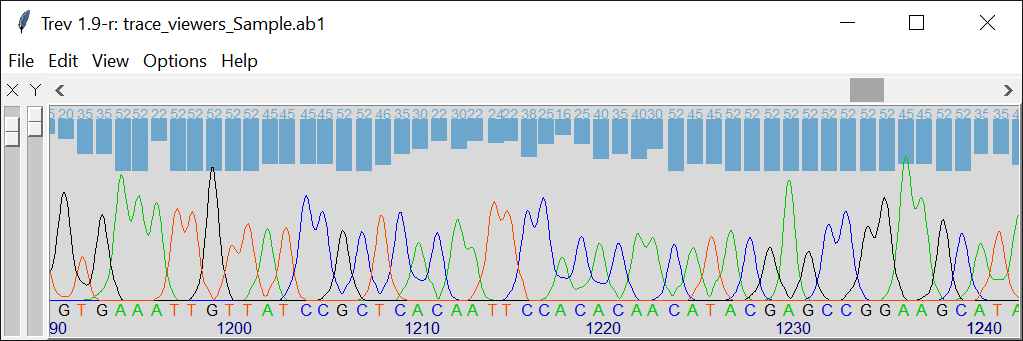
Trev Trace Viewer
Developer: Medical Research Council (MRC) Laboratory of Molecular Biology, Cambridge, England.
Supported platforms: Unix, Linux, MacOSX and MS Windows.
Download: http://staden.sourceforge.net/
Features
View Quality scores:
Yes. The quality scores can be toggled on or off and are displayed above the peaks of the chromatogram as bars and values.
View raw data:
No.
Assembly:
Within the Staden package there are other tools for sequence assembly, such as Gap 4 and 5 but not within Trev trace viewer.
Input file formats:
ABI, ALF, SCF, ZTR, CTF, Plain ASCII txt, Experiment (staden).
Output file formats:
SCF, CTF, ZTR, Experiment and plain txt.
Trimming:
No.
Integrated Blast:
No.
Open multiple windows:
Yes.
Edit bases:
Yes. A second sequence identical to the sequence is generated and that sequence can be edited and exported.
ABI limits (regions outside of clear range region are displayed in gray):
No.
Other functions
you have the ability to manually shade the 5′ and 3′ ends of a sequence to perform a soft trimming of unwanted areas.
Cost
The Staden Package software that includes Trev trace viewer is free.
Comments
Trev trace viewer feels dated and lacks many features that other available trace viewing applications have. The quality bars and peaks can overlap which can be confusing.