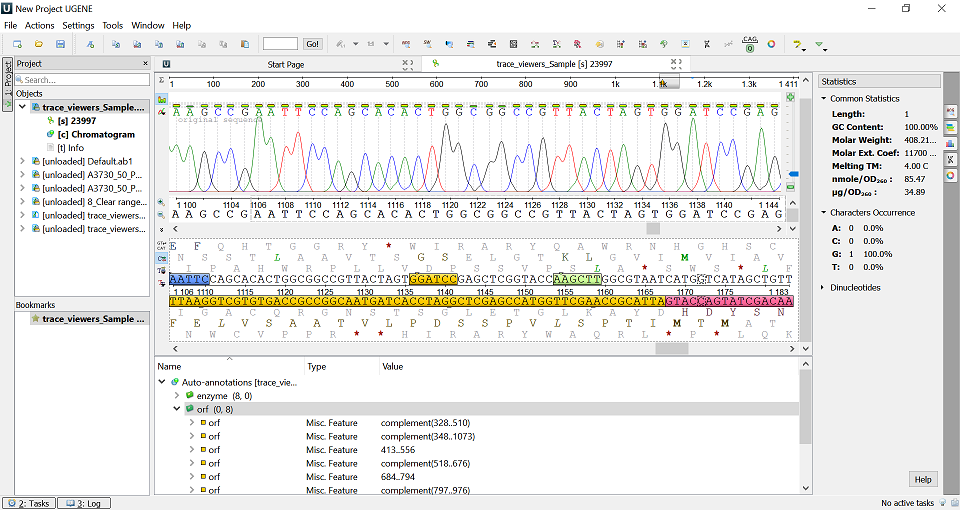
UGENE main window view
Developer: Uniporo
Supported platforms: Windows XP, Vista 7/8, Solaris 10, Linux and OS X
Download: UGENE Downloads
Features
View Quality scores
Quality scores are available above each basecall as colored bars. However they can be quite hard to read.
View raw data
There is no way to view the raw data through UGENE.
Assembly
Assembly is available in UGENE through the CAP3 plugin
Input file formats
.abi and .scf
Output file formats
.scf, FASTA, FASTQ, Genbank, GFF and Vector NTI Sequence
Integrated Blast
Yes, UGENE has supported full control over NCBI’s BLAST remote and local databases.
Open multiple windows
yes, you can have multiple project windows open at the same time with the ability to have multiple sequences in each project.
Edit bases
Yes.
ABI limits (regions outside of clear range region are displayed in gray):
No.
Other functions
UGENE is a bioinformatics workhorse, there are multiple tools and plugins available to do all your sequence annotation and assembly work. These tools include but are not limited to, ORF, reapeat, tandem, HMM and tanscription-factor binding sites finder, plasmid annotation, reverse compliment sequence, restriction sites and multiple graphical interfaces.
Cost
UGENE is free and open sourced under a GNU GPL2.
Comments
Although UGENE provides all the tools a scientist would \need to analyse and assemble DNA sequences, you can’t help but feel that you need a 2 day crash course to know how every function works. The manual is nearly 300 pages long and it can take a while to find out how to do what you want to do. If you are looking for a simple trace viewer this isn’t for you, however if you want to do analysis of your trace this will serve you well, just give yourself a couple of hours to familiarize yourself with the program.