CounterTrace II™ DNA sequencing system is an upgrade of the CounterTrace DNA sequencing system for use with the ABI 3730 KB base caller. The CounterTrace II sequencing system offers the following features for scientists sequencing DNA:
- Up to 30% more KB Q20 bases per trace
- Compatibility with the ABI 3730 and 3730xl DNA sequencing instrument
- Ease of use in both large and small scale DNA sequencing facilities
Overview of CounterTrace II software & reagent
Current state-of-the-art capillary DNA sequencers can produce reads that can often exceed 1000 bases. However, these long dideoxy sequencing reads typically suffer from excessive peak broadening and poor resolution in the later part of the trace. This latter region is associated with low quality scores and high base call error rates.
CounterTrace II uses an internal calibration standard to allow the accurate measurement and processing of each sequencing trace (Figure 1). The CounterTrace additive contains DNA fragments of known size labeled with a proprietary fluorophore (CounterTrace standard). This fluorophore has been designed to enable its specific detection in four channel sequence data. Addition of the CounterTrace standard to each sequencing reaction after sample clean-up provides unique ‘landmark’ sizing peaks in the raw trace data. The CounterTrace II software uses the location of the standard peaks to enable accurate sizing and processing of the trace data. After algorithmic removal of the standard peaks from the trace data, the CounterTrace II software output sequencing trace file in the ab1 format. These trace file are then able to base- and quality-called using the KB DNA sequencing base caller.

Figure 1. Overview of the CounterTrace II DNA sequencing process.
Benefits & features of the CounterTrace II DNA sequencing system
Easy implementation for DNA sequencing facilities
The CounterTrace II system is simple to implement into any DNA sequencing facility. The CounterTrace additive is diluted to the appropriate concentration in the sample loading solution used by the facility (e.g. water, 50 µM EDTA, or formamide) and the sequencing sample processed as normal. After collection of the trace data, the CounterTrace II software is used to reprocess the traces before base and quality calling with KB v1.1. The CounterTrace II DNA software includes an automation feature that allows seamless integration into existing data pipelines.
CounterTrace II is compatible with the ABI BigDye™ v1.1 and v3.1 sequencing chemistries using the ABI3730 and ABI 3730xl DNA sequencing instruments for both the 36cm and 50cm capillary array lengths.
Increase in DNA sequencing trace read length
CounterTrace II allows the poorly resolved regions found at the end of the DNA sequencing trace to be accurately processed, thus providing the user with significantly longer and higher quality reads (Figure 2 & 3).
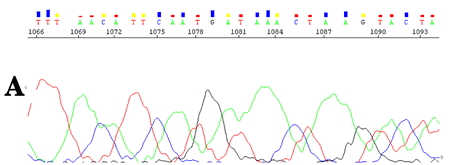
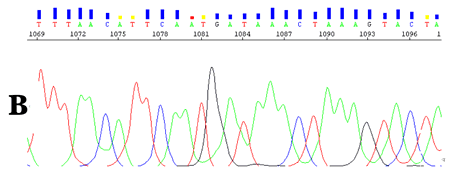
Figure 2. KB v1.1.1 derived base and quality calls from an ABI3730xl 50 cm capillary array DNA sequence trace. A. Before CounterTrace II processing. B. After CounterTrace II processing.
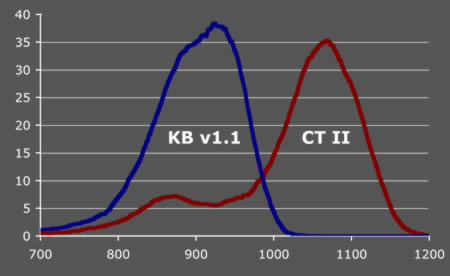
Figure 3. Distribution of number of Q20+ bases per trace before (KBv1.1) and after CounterTrace II processing (CT II). Results from 5427 traces generated on an ABI3730xl 50cm capillary array.
CounterTrace II offers increases in alignable KB Q20+ read lengths of up to 30% for individual DNA sequencing traces, with an average of 15–20% in high throughput production environments.
Increased automated DNA sequencing throughput
CounterTrace II processed traces run on the ABI3730 36cm capillaries provided DNA sequencing read lengths previously observed only with 50cm arrays. As these short-array runs take only half the time required for the longer arrays, the throughput can be effectively doubled by using CounterTrace II without the capital cost involved in purchasing additional instruments. In addition, Nucleics offers a modified 36 cm array run module that enables up to 28 runs per day (~51 min run time) to be performed without sacrifice of sequence read length.
The improved DNA sequencing read lengths provided by CounterTrace II increases the throughput and lowers the overall per base cost. In addition, these longer read lengths enable DNA sequencing projects (e.g. whole genome, cDNA clones, etc) to be completed at lower sequence coverage levels, thus offering further cost- and time-savings.
The CounterTrace II DNA sequencing software is available for Windows 2000, XP and x86 Linux. For addition information on the CounterTrace II DNA sequencing system please see the CounterTrace II DNA sequencing Quick Start guide, or contact our support team with any of your technical questions.
CounterTrace II Price & Ordering Information
CounterTrace II has been superseded by our PeakTrace basecallers. While CounterTrace II will continued to be made available to existing users we strongly recommend that they email us at {This email is obscured. Your must have javascript enabled to see it} to obtain information on upgrading to PeakTrace.