Introduction
The Auto PeakTrace RP™ DNA sequencing software extracts, processes and basecalls the raw data contained within ABI and MegaBACE trace file to discover additional high quality bases. This data reprocessing enables Auto PeakTrace RP to make significantly better base and quality calls than can be achieved using the KB Basecaller™, or other basecallers such as phred or TraceTuner. The PeakTrace software works with .ab1, .abi and .abd files generated by the ABI 377, ABI 310, ABI 3700, ABI 3100, ABI 3130, ABI 3730, ABI 3730xl, ABI 3500, ABI 3500xl, MegaBACE 1000, MegaBACE 4000, and the MJ Research BioStation DNA sequencing instruments.
The Auto PeakTrace RP software is able to provide a completely seamless and straightforward method to increase DNA sequence read length without needing to change your current DNA sequencing production protocols.
PeakTrace basecalling can be performed any time after the trace data is collected. This allows it to be used with previously collected sequencing data sets such as partial genome sequencing projects. By applying PeakTrace basecalling on trace data from incomplete sequencing project and reassembling the resulting DNA sequences, the overall genome assembly can be significantly improved without the need to collect more sequence data. Since PeakTrace can output trace files in the ABI .ab1 format, it is compatible with other software applications requiring files in this format.
Auto PeakTrace RP performs remote PeakTrace processing of trace files without the need and expense of a PeakTrace:Box™ system. Auto PeakTrace RP offers the same control over the processing parameters that the PeakTrace:Box system does with the exception of TraceTuner basecalling and QualTrace III.
Announcements of updates to Auto PeakTrace RP are posted to the Nucleics Twitter (@nucleics_inc) and Nucleics Facebook accounts.
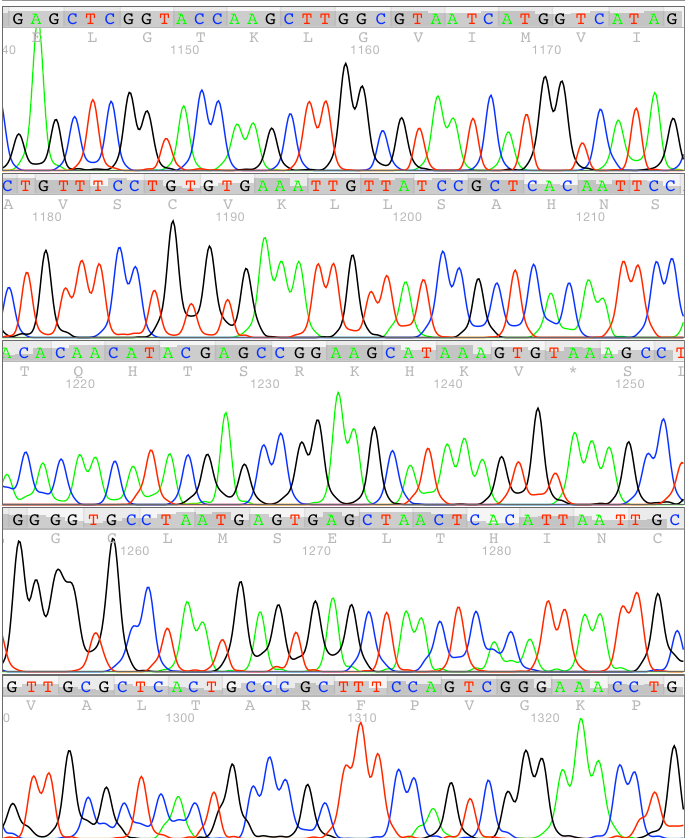
An example of what Auto PeakTrace RP can achieve can be seen below. This is a normal ABI 3730 50cm array trace file showing the regions 1140 to 1330!
How to buy or upgrade a license
To purchase a license for Auto PeakTrace RP, or to add units to your current license, please contact Nucleics at {This email is obscured. Your must have javascript enabled to see it}.
In China please contact our exclusive agent Shanghai BOQ.
Auto PeakTrace RP Release History
6.23-24
- Improvements to increase the robustness of the software on very fast networks.
- Improved cleaning of temporary files from the Application Data folder.
- Upgrade of cURL to 7.48.0.
- Other minor bug fixes and enhancements.
6.22
- New Feature. User settable PCR trim base.
- All N bases now have a quality score of 2 rather than 0.
- Improved handling of licensing issues.
- Bug fixes and enhancements.
6.21
- New Feature. Additional Australian and Chinese based RP server.
- More accurate estimated time to completion function.
- Improved handling of proxy server issues.
- Bug fixes.
6.20
- New Feature. Regional based servers to improve processing speed. Use of the new servers may require a new license code.
- New Feature. User control over file upload encryption. Turning off encryption can speed the upload speed.
- Dynamic speed adjustment to match upload speed with network speed. This should result in much faster trace processing on fast networks.
- Bug fixes and internal improvements to improve robustness.
6.12
- New Feature. Bubble spike removal.
- Fixed a bug preventing a minus value being manually entered for good base improvement.
- Removal of the command line version (peaktrace.exe) from the Auto PeakTrace 6 Installer.
- Other internal bug fixes.
6.10
- New Feature. User controlled extra smoothing of traces.
- New Feature. Manual control of the skip short base threshold.
- New Feature. Limit trimming to the 3′ end of the trace only.
- New Feature. Set use of the edited basecalls within the trace.
- New Feature. Set the output to overwrite the input traces.
- New Feature. Picket Fence trace output.
- New Feature. PeakTrace basecalling with KB fall back.
- New Feature. Simple mixed basecalling.
- A reorganized and simplified Options window.
- Improved fuse trace of repeat sequence regions.
- Improved fuse basecall of repeat sequence regions.
- Improved basecalling of mixed peaks.
- Bug fixes.
6.00
- New Feature. Clean baseline.
- New Feature. Extra normalization.
- Parallel installation of Auto PeakTrace 6 RP with Auto PeakTrace RP.
- Improved handling of capillaries that run slower than expected.
- Improved signal to noise determination.
- Improved trimming of noise level data from traces.
- Improved detection of failed traces.
- Improved basecalling of traces without KB quality scores.
- Improved basecalling of short traces.
- Improved q average trim.
- Additional error messages to help identify the cause of problems.
- Updated visual design and help manual.
- Bug fixes.
5.90
- New Feature. Support added for the following run conditions:
- ABI 3500 Ultra Rapid Run POP 7 BigDye 3
- ABI 3500 Ultra Rapid Run POP7 BigDye 1
- ABI 3500 POP 6 BigDye 3
- ABI 3500 Ultra Rapid Run POP 6 BigDye 3
- ABI 3500 Ultra Rapid Run POP 6 BigDye 1
- ABI 3100/3730 POP 7 BigDye 1 (11 to 12.5 kV)
- ABI 3100/3730 POP 7 BigDye 1 (11 to 12.5 kV)
- ABI 3100/3730 POP 4 Rapid BigDye 3
- ABI 3100/3730 POP 4 Rapid BigDye 1
- ABI 310 30cm array POP 4 Rapid BigDye 1
- ABI 310 30cm array POP 6 Rapid BigDye 3
- ABI 310 30cm array POP 6 Rapid BigDye 1
- ABI 310 36cm array POP 6 Rapid BigDye 3
- ABI 310 36cm array POP 6 Rapid BigDye 1
- ABI 310 50cm array POP 6 Rapid BigDye 3
- ABI 3730 36cm array POP 7 ET
- New Feature. New error condition: No raw peak data
- Improved support for ABI 310, ABI 3500 and MegaBACE traces.
- Improved basecalling and trace processing.
- Many bug fixes and improvements.
5.83
- Bug fixes.
5.82
- Additional error messages and warning to aid use.
- Improved processing speed.
- 7za.exe updated to 9.38
- Bug fixes.
5.81
- Improved robustness when using slow or unreliable networks and/or remote shared drives.
- Bug fixes and help manual updates.
5.70
- New Feature. Auto selection of the best first peak location.
- Improved fusing of traces containing large repeat regions.
- Improved trimming of low quality bases from trace ends.
- Improved basecalling of mixed signal traces.
- Improved detection of duplicate files in the input folder.
- Improved processing of mixed signal traces.
- Improved error messages.
- Bug fixes.
5.61
- Trace files are now encrypted while being transfered to and from the remote PeakTrace server.
- Improved transfer speeds (~10%) due to improved compression of trace data.
- Greater robustness to slow or intermittent network connections.
- Additional error messages.
- The usual bug fixes.
5.60
- New Feature. Failed traces are now not charged. The exceptions are:
- Traces below the good base improvement threshold where the good base improvement threshold is set to a value higher than -10. If using the default value of -20 you will not be charged.
- Traces that can’t be output because the output folder is unwritable.
- New Feature. Support added for the following run conditions:
- ABI 3730 Low Voltage POP7 BigDye 3.1
- ABI Limits are only updated for successful traces
- Improved basecalling and trace trimming.
- The usual bug fixes and enhancements.
5.50 -5.51
- Improved fuse basecalling option.
- Improved basecalling through large dye peaks.
- Default options changed to improve basic usage
- .seq file output: abi format
- required good base improvement: -20
- q average trim: Q12 average in 20 base window
- n base threshold: 5
- skip short traces: yes
- A short trace is now considered to be a trace with fewer than 500 bases.
- Improved support for ET terminator traces.
- Improved support for ABI 3500 BigDye 1.1 traces.
- New error code 36 added – Input and output paths are the same.
- New error code 38 added – Skipped short trace.
- Good quality threshold and N base threshold now have maximum values of 99.
- The usual bug fixes and tweaks.
5.41 – 5.42
- Bug fix for problem with automatically extracting proxy server settings.
- Chinese language version.
5.40
- New Feature. Unicode path support.
- New feature. Unicode logfile output.
- UI improvements.
- Most likely some new bugs introduced due the large number of changes (~3000) needed to support Unicode paths 🙁
5.32
- New Feature. Check for corrupt settings. If the settings file is corrupt the settings are reset to the factory default.
- Simplified EULA.
- Bug fixes and UI enhancements.
5.30 – 5.31
- Bug fixes.
5.20
- New Feature. Support added for the following run conditions:
- ABI 3130 Rapid Run 50cm Array POP6 ET Terminator
- ABI 3130 Rapid Run 36cm Array POP4 BigDye 1.1
- ABI 3730 Ultra Low Voltage POP7 BigDye 3.1
- New Feature. Archives input traces after processing traces.
- New Feature. 15% faster processing.
- New Feature. Folder browsing starts from last used folder.
- New Feature. Additional remote server added located in Eastern USA.
- Improvements to forced processing to enable more traces to be processed.
- Numerous bug fixes.
5.13
- New Feature. Option settings also saved to a plain text file when Auto PeakTrace RP is quit.
- Improved error messages.
- New folder browser dialog.
- Bug fixes.
5.12
- Improved error messages.
- New folder browser dialog.
- Better management of processing rates based on Internet speed.
5.11
- Improved alignment of early basecall to peaks when using fuse basecall option.
5.10
- New Feature. Fuse Basecall.
- New Feature. No Peak Resolution.
- Multiple peaks are now no longer allowed to be located at the same data point in the trace.
- Warning provided when less than 100 units remain on account.
- Bug fixes.
5.09
- Wine-based MacOS X version released.
- Improved server connection error reporting.
- Bug fixes.
5.05 – 5.08
- New Feature. Support for additional ABI 3500 run types.
- Bug fixes.
5.04
- Updated license conditions.
- Improved trace processing.
- Automatic restart upon program crash.
5.03
- New Feature. Added support for abi+kb trace output.
- Add a warning for input and output paths contains non-ANSI characters.
- Minor bug fixes.
5.02
- First public release of Auto PeakTrace RP.
System Requirements
The minimum recommended system configurations for Auto PeakTrace RP
- Windows XP, Windows Vista, Windows 7, Windows 8, or MacOS X 10.6+
- Functioning Internet connection
- 512MB of RAM
- 100MB of free disk space
- 1024 x 768 screen resolution or higher
The limiting factor for processing speed in most cases is your Internet speed.
Installing Auto PeakTrace RP
Windows
To install Auto PeakTrace RP just double click the Auto PeakTrace RP Installer icon. You will be prompted through the installation process by the Auto PeakTrace RP installer.
To remove Auto PeakTrace RP select the Uninstall menu option from the Auto PeakTrace RP folder in the Windows Start menu.
To purchase units select the Purchase Units option from the Auto PeakTrace RP folder in the Windows Start menu.
Mac OS X
A version of Auto PeakTrace RP is available that will run on MacOS X under Wine. To install Auto PeakTrace RP open the Auto PeakTrace RP Installer.dmg file and drag and drop the Auto PeakTrace RP.app file as instructed to your Applications folder. Auto PeakTrace RP can be run like any other Mac OS X application, however, the application will look like a Windows XP program.
To remove Auto PeakTrace RP just drag and drop to the application to the Trash.
Warning
While it is possible to install two versions of Auto PeakTrace RP at the same time, attempting to run two versions at the same time will cause each version to corrupt the settings of each version (both versions will share the same options file). If you want to run an older version of Auto PeakTrace RP after installing a new version please remember to reset the options back to their defaults before using.
Getting Help
If you are having any problems Nucleics provides technical support for the Auto PeakTrace RP software.
Please address any questions or comments to {This email is obscured. Your must have javascript enabled to see it} or visit the Auto PeakTrace Forum.
Quick Start Guide
To run the Auto PeakTrace RP software either double click the desktop icon or select Auto PeakTrace RP from Windows Start menu. Your computer will need to have a functioning internet connection for the processing of traces to proceed.
Auto PeakTrace RP has a single window. Optional processing parameters can be accessed via the Options button. Help can be obtained using the Help button.
License codes can be entered via the License Code window by clicking on the License Code Button from the Option window. Licenses can be obtained by contacting Nucleics.
To purchase units select the Purchase Units option from the Auto PeakTrace RP folder in the Windows Start menu.
To run Auto PeakTrace RP choose the Input and Output folders. The Input folder should contain either .ab1 files or folders of .ab1 files. The Output folder should be the location that you wish to save the PeakTrace basecalled files. The Input and Output folders can be selected by file browsing to the appropriate locations using the Browse buttons located on the left of the Main Window.
PeakTrace basecalling is started by pressing the Run button.
The Auto PeakTrace RP software checks for any new trace files found in Input folder at the frequency set by the check for new files setting in Options window. Any new traces files, or folders of trace files, that are found in the Input folder are basecalled and the results saved to the Output folder.
Auto PeakTrace RP processing can be stopped by pressing the Stop button, or when all files have been processed.
Using Auto PeakTraces RP

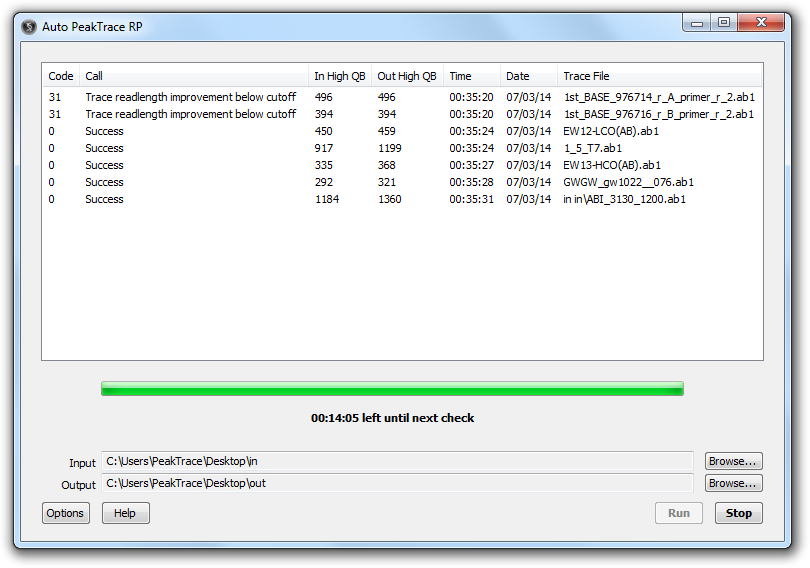
Results Window
This shows the output results of the PeakTrace basecalling. The following results are shown for each trace file.
- Code. A numerical code of basecalling result.
- Call. A short description of basecalling result.
- In High QB. The number of high quality bases in the input file.
- Out High QB. The number of high quality bases in the PeakTrace basecalled file.
- Time. Time of PeakTrace basecalling.
- Date. Date of PeakTrace basecalling
- Trace File. The file name of the input files including any sub-folders.
Input
The name and full path to folder containing the .ab1, .abi or .abd trace files you wish to basecall. This folder can be on your computer, or any computer accessible by Windows File Share network (e.g. the ABI DNA sequencer). All traces that you want to basecalll need to be located in this folder, or in sub-folders within this folder.
Output
The folder where the PeakTrace basecalled trace file and other associated files are saved. The files will be saved using a folder structure that mirrors the input folder. For example, if the input files are all located in a sub-folder called sub123, then the output files will be saved to the sub-folder sub123 in the output folder.
Also written to the Output folder is a tab delimited log file summarizing the basecall results for each trace collection. The log file is called peaktrace.log and contains the same columns as shown in the Results Window. A Q20+ base are any basecalls that PeakTrace predicts have quality score of 20 or higher. A quality score of 20 corresponds to an expected base call error rate of 1 in 100.
Progress Bar
Provides an estimate of the progress of the PeakTrace basecalling. Underneath the progress bar, the percentage of traces that have been processed, the number of traces that have been improved, and the estimated time remaining is shown. Once the processing has finished a countdown to the next check will then be shown.
Run
Starts the basecalling of trace files by PeakTrace. The first check will be done immediately. The second check for new trace files occurs after the elapsed time selected in Options. The default wait time is 15 minutes.
Stop
Stops the basecalling of new trace files. Pressing the Run button will restart the basecalling.
Options
See Options section for details of the various PeakTrace procession options.
Help
Displays the Auto PeakTrace RP Help Manual.
Options Window
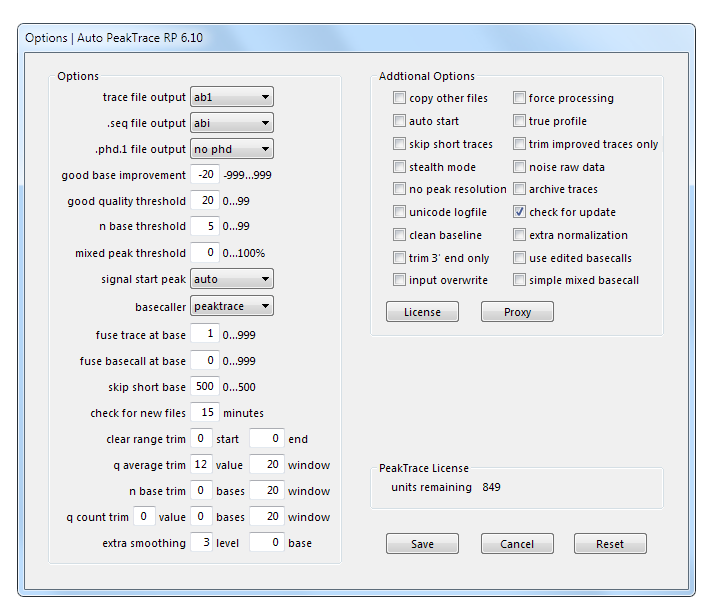
Auto PeakTrace RP Options Window.
All Auto PeakTrace RP basecalling and processing settings are accessed via the Options button. To save any changes press the Save button. To cancel any changes press the Cancel button. To revert to the original default settings press the Reset button.
PeakTrace Options
These setting allows full control over the PeakTrace basecalling, file output and formating.
trace file output
Sets the output trace file format. The options are ab1, ab1+scf, ab1+kb or scf format. The .ab1 file format is only available for trace files obtained from ABI 3730, ABI 3500, ABI 3130, or ABI 3100 DNA sequencers. The ab1+scf option outputs trace files in both .ab1 and .scf format. The ab1+kb outputs traces file in .ab1 format and also copies the original KB basecalled trace to the output folder appending _kb to the trace filename. This option is useful if you wish to supply end users both the PeakTrace basecalled trace file and the original KB basecalled file. Default is ab1.
.seq file output
Sets the output of text based .seq basecall files. The format options are abi, fasta, fasta+qual or noseq (no .seq file). The fasta+qual option generates two files, a .seq file in fasta format and a .qual quality score file. Default is abi.
.phd.1 file output
Sets output of .phd.1 format files. The options are phd or nophd (no .phd.1 file generated). Default is nophd.
good base improvement
The minimum number of high quality bases that PeakTrace must improve the basecall to generate the trace file. The allowed values can range from -999 to 999 bases. If the PeakTrace basecalling does not generate more high quality bases than this value then the original input trace file is copied to the output folder. Default value is -20 bases.
good quality threshold
The quality score threshold that is used for a base to be considered a high quality base. The allowed values can range from 0 to 55. Default value is 20.
n base threshold
The minimum quality score threshold for basecalling. Any basecalls with a quality score equal to, or below, this threshold value are set to a N base. Default is 5.
mixed peak threshold
The threshold for secondary peaks found under the primary peak to be basecalled as mixed (degenerate) bases. Default is 0 (no mixed basecalling performed).
signal start peak
Sets the location of the first peak used for basecalling. If first is selected the original peak location generated by the ABI data collection software is used. If second is selected, the most recent KB basecall peak location is used. This setting is useful if you wish to change the start point at which basecalling begins. Default is auto.
basecaller
Sets the basecaller used by PeakTrace. For most applications you will want to use the PeakTrace basecaller (peaktrace). An alternative is to output to the original KB basecall using the abi option. The abi option is useful if you wish to convert .ab1 traces files to .scf format, or generate the other sequencing files from the KB basecall (i.e. .seq and .phd.1 files). Default is peaktrace
fuse trace at base
This option causes the PeakTrace output trace to be fused with original KB basecalled trace. This setting allows the better KB basecalling through dye blobs (large dye peaks) to be combined with better traces processing of PeakTrace at the 3′ end of the trace. The point at which this fusion can be set is between base 0 and 1000. It is recommended that you use as low as value as possible (1 is good), but if you have significant late dye blobs you may want to use a higher number to set the fuse point past the trace region affected by these large dye peaks. It is highly recommended to use the default setting with all KB basecalled traces. Default is 1 (on).
fuse basecall at base
This option causes the PeakTrace basecall to be fused with the original KB basecall using the PeakTrace processed peaks. The basecall fusion can be set between base 0 and base 200. This option is is an alternative to using the fuse trace option. If both fuse trace and fuse basecall are set the fuse trace setting is used and the fuse basecall option ignored. It is recommended that you use as low as value as possible (1 is normally best).
Due to inherent difficulties in aligning basecalls to peaks in the very early trace region, this setting may result in peak location shifts in the first 40 bases. This can cause problems when combined with mixed basecalling as the peak locations may not correspond to the basecall. For this reason the fuse trace option is recommended over the use of this setting. Default is 0 (off).
check for new files
The time interval that Auto PeakTrace waits to checks for new trace files in the Input folder or sub-folders. This can be used to automatically process trace files as the DNA sequencer generates them. Default is 15 minutes.
clear range trim
Sets the clear range clipping range. Bases before start or after end are removed from the 5′ and 3′ ends of the trace file and associated sequence files. For example, setting the range from 20 and 800 removes bases 0-19 and 800+ from the trace. Default is no clear range trim (0 and 0).
q average trim
Sets the quality score average clipping threshold and window size. The traces are clipped using a sliding window of defined size when the average quality score falls below the set q value. For example, if value is set to 10 and window to 20, this option will remove bases from 5′ and 3′ ends where the average sliding 20 base quality is below 10. This setting is useful for removing low quality regions from PCR product reads. Default is q score trim of 12 and 20).
n base trim
Sets the N base score clipping threshold and window size. The traces are clipped using a sliding window of selected size that has more than the selected number of N bases. For example, if bases is set to 10 and window to 20, this option will remove bases from the 5′ and 3′ ends of the sequence where the sliding window of 20 base has more than 10 N basecalls. Default is no n base trim (0 and 20).
q count trim
Sets the quality score count clipping threshold, number of bases and window size. The traces are clipped using a sliding window of the selected size that has more than the selected number of bases with a quality score below the quality score value. For example, if value is set to 20, bases is set to 4 and window to 20, this option will remove bases from the 5′ and 3′ ends of the sequence where a sliding window of 20 base has more than 4 basecalls with a quality score below 20. Default is no q count trim (0, 0 and 20).
copy other files
Copies all non-.ab1 files found in the Input folder to the Output folder. This option is useful if you wish to mirror the contents of Input to Output. The option can cause problems if you have .phd.1 or .seq files generated by the KB basecaller located in the Input folder and also select to output .seq or .phd.1 files using PeakTrace. Default is unchecked (off).
force processing
Forces basecalling of trace files that are normally rejected by PeakTrace due to poor quality. Use this option with care as force processing can result in the generation of corrupt and/or nonsense files. Default is unchecked (off).
auto start
Start the automated processing of trace files once Auto PeakTrace RP is launched. Requires that the Input and Output folders have been select and saved. This option is useful if you wish to have Auto PeakTrace RP automatically run after restarting your machine. Default is unchecked (off).
true profile
Outputs traces using the true peak signal intensity. Default is unchecked (off).
skip short traces
Does not process or count usage for traces with less than 500 bases. This setting is useful if processing data sets that contain short PCR products. Default is checked (on).
trim improved traces only
Applies base trimming to the PeakTrace improved traces only. Default unchecked (off).
stealth mode
Hides that the trace was basecalled by PeakTrace. Both the .phd.1 and .scf file will record the trace processor and basecaller are KB 1.4. Default is unchecked (off).
noise raw data
This replaces the raw data channels with random noise. This can be used to prevent basecalling of the resulting .ab1 file by the KB basecaller. Default is unchecked (off).
no peak resolution
This provides a trace output where overlapping or broad peaks are not resolved in the trace file and where the signal intensity is proportional to the raw data signal intensity with a basecalling quality equivalent to normal PeakTrace processing. This option is designed for the detection of polymorphisms (double peaks), or for applications where the superior basecalling of PeakTrace is desired, but where the peak intensity is proportional to the raw signal intensity. Default is unchecked (off).
archive traces
This option moves the input trace files into a sub-folder of the input folder called archive after PeakTrace processing. This prevents the input traces from being processed again if run is pressed before the old files are removed. Default is unchecked (off).
unicode logfile
This option outputs the logfile as Unicode rather than ASCII text. This allows non-English characters to be displayed in the logfile. Because this can cause downstream applications to break it is recommend that you don’t use Unicode logfiles unless you require this option. If you choose the default ASCII encoding any non-ASCII characters will be displayed as “?”. Default is unchecked (ASCII encoding).
license code
This button lets you to enter your Auto PeakTrace RP license code.
proxy settings
This button allows the proxy server setting to be manually modified. Only use this if you are having trouble connecting to the PeakTrace server.
License Usage
The units remaining on your license are shown below the main Options settings. This may be blank until you first process a set of trace files.
If your remaining units are getting low (less than 3 days typical usage) then please contact Nucleics to add more units to your account.
Proxy Settings Window
If your internet connection is not working the most likely cause is that you are behind a proxy server (very common in corporate and university environments). Auto PeakTrace RP will attempt to automatically workout your proxy settings, but if you have an unusual proxy setup it may require that you manually enter your proxy server details.
The proxy settings are accessed from the options window by clicking on the proxy settings button.
Enter the proxy server details and port number. If you do not know these then please contact your IT department.
It is possible that your proxy server may require a username and password. Most proxy servers don’t require these, but if you are still having trouble connecting after entering the proxy server details then check with your IT department to see if you need a proxy server username and password to access the internet.
License Code Window
Enter your license code that you have obtained from Nucleics. To purchase a license please see how to buy a license. Please be careful entering the code as entering the incorrect code will prevent Auto PeakTrace RP from being able to connect to your account on the PeakTrace server.
Auto PeakTrace RP checks the license code for typing errors so if it suggests that the code has a typing error check very carefully what you have entered.
PeakTrace Basecalling Results
Auto PeakTrace RP reports on the result of PeakTrace basecalling as well as any reason for not being able to basecall a trace file.
success
Trace was processed and basecalled by PeakTrace with the total number of high quality bases exceeding the good base improvement threshold.
trace readlength improvement below cutoff
The improvement in readlength are less than the good base improvement threshold selected in Options. The output file is the original .ab1 file. This prevents the generation of traces with reduced readlength if PeakTrace is not able to improve the basecall. If this error is occurring with good quality traces then try resetting the options values to the default values using the Reset button and processing again.
weak or failed trace
Trace file contains no or only very weak peak signal in the raw data channels, especially after base 600. This result can be caused by:
- No DNA template.
- Loss of sample during clean up.
- A blocked capillary in the array
- Heavy contamination of the template DNA with salt or other charged ions.
Alignable Q20+ bases are typically low.
mixed or noisy raw peaks
Trace file contains a significant number of double peaks. This result can be caused by the presence of:
- Two different DNA templates.
- Two primer binding sites.
- Primer mis-annealing at secondary sites.
- Single primer PCR amplification in the sequencing reaction.
Alignable Q20+ bases are typically low or moderate.
late peak signal at noise level
Trace file has a very low signal-to-noise ratio (below 4) before base 700 for 36cm capillary arrays traces, or base 900 for 50cm capillary array traces. Very low signal-to-noise ratios severely limits the sequence read length improvement possible by PeakTrace. This result has many causes including:
- Incorrect template concentration.
- Excessive sequencing chemistry dilution.
- DNA sequencing instrument failure.
- Blocked capillaries.
- Contamination with salts, detergents, proteins, RNA, or excessive DNA.
Alignable Q20+ bases are typically moderate to high.
unsupported run condition
PeakTrace does not support the DNA sequencer run conditions used to generate the trace file. This result can occur if you are using custom run conditions, or because of an unusually combinations of run voltage, polymer type, instrument, dye chemistry, and/or capillary lengths. If you obtain this error please contact Nucleics as we can add support for new run conditions relatively quickly once we have some example trace files. The run conditions that are currently supported are listed here.
PeakTrace Log file Format
The PeakTrace log file is tab delimited with the first line a header. A typical example of the peaktrace.log file is shown below along with a description of what each column means.
Code Call In High QB Out High QB Time [hh:mm:ss] Date [mm/dd/yy] Output Path 0 Success 830 913 08:41:46 02/07/08 C:\temp2out\101-_D08_Seq121406p4.ab1 0 Success 1184 1316 08:41:49 02/07/08 C:\temp2out\3130_1200.ab1 0 Success 1156 1316 08:41:51 02/07/08 C:\temp2out\3130_1500.ab1 0 Success 795 851 08:41:53 02/07/08 C:\temp2out\681+_E10_Seq120606p4.ab1
Code
A numerical code of the basecalling result. The following codes can be generated by Auto PeakTrace RP.
0 Success 2, 36, 37, 38 Can't open input trace 3, 40 Can't create output trace 5 Can't write output trace 10 No processed peak data 12 Unsupported run condition 1, 4, 14, 17, 23 Out of range 15 Late peak signal at noise level 19 Spacing model failure 20, 26, 39 Bad command line arguments 27 Weak or failed trace 31 Trace readlength improvement below cutoff 32 Basecalling failed 33 PeakTrace license expired 34 Processing exceed limit 36 Input and output paths are the same 38 Skipped short trace 35, 41 - 45 Cannot connect to remote server
The same error call can be generate different error codes. These codes are useful for diagnosing the cause of the error.
Call
A short description of the basecall result.
In High QB
The number of high quality bases found in the input trace.
Out High QB
The number of high quality bases found in the output trace.
Time
The PeakTrace run time in hours:minutes:seconds format.
Date
The PeakTrace run date in month/day/year format.
Output Path
The output path and file name of the output trace file.
Auto PeakTrace RP Errors
The following errors may occur when attempting to basecall trace files. The possible causes of these errors and their corresponding code are described, together with possible solutions. In most cases these reflect corrupt trace files or program malfunctions.
can’t open input trace
This error may occur if the file does not exists or if the file has incorrect access permission settings. Check if file exists and that you have read permission to the Input folder. Incorrect access permissions can occur if Windows or PeakTrace crashes.
can’t create output trace
This error may occur if you do not have write permission to the Output folder, or if the drive is full. If the drive is full please remove excess files and restart Auto PeakTrace RP.
can’t write output trace
This error may occur if you do not have write permission to the selected Output folder, or if the drive is full. Please remove excess files and check if you have write permissions with the Output folder selected.
no processed peak data
This error may occur if the trace file is corrupt or has not yet been basecalled by the KB or ABI basecaller.
basecalling failed
This can occur when poor quality traces are force processed.
out of range
This error indicates the trace data lies outside the expect model for the run condition used. These traces are very useful for improving PeakTrace so if you obtain this error we would appreciate if you could share your data with us so that we can continue to improve PeakTrace.
spacing model failure
This error occurs if PeakTrace is not able to measure the average peak spacing from the raw data. This can occur with poor quality traces with mixed or very weak signal.
processing exceed limit
This error occurs if the trace cannot be basecalled within 20 seconds. This error indicates a program malfunction or a very slow internet connection. If your internet connection is fine please retain the traces file that provides this output and contact Nucleics.
peaktrace license expired
Indicates that you have used all your licensed number of trace processings. Please contact Nucleics to obtain a license upgrade.
cannot connect to remote server
Indicates that Auto PeakTrace RP can’t connect to the remote PeakTrace server. This can occur because your Internet connection is not functioning, the proxy server details are missing or incorrect, or because you have entered the wrong license code. Please see the troubleshooting guide for suggestions on how to solve this error.
Troubleshooting Guide
First check that you have a functioning internet connect and can connect to the RP server (rp.peaktrace.com) using a browser.
If your internet connection is functional, but Auto PeakTrace RP is still not working, the most likely problem is that you are behind a proxy server (very common in corporate and university settings). Auto PeakTrace RP will attempt to automatically discover your proxy settings, but if you have an unusual proxy setup it may require that you manually enter the proxy settings.
The proxy settings are accessed from the options window by clicking on the proxy settings button.
Enter the proxy server details and port number. If you do not know these then contact your IT department.
It is possible that your proxy server may require a username and password. Most proxy servers don’t require these, but if you are still having trouble connecting then check with your IT department to see if you need a proxy server username and password to access the internet.
No License Entered
Before you can process any traces you need to enter your license code (a 9 or 10 character long value) into the License Code Window. Do not enter any spaces.
Unable to Connect to Account on RP Server
The most common cause of this error is entering an invalid license code. This error can also occur if the PeakTrace server and the secondary server are off-line or your account is inactive. If you have checked your license code carefully and it is correct please contact us so we can check if the server or your account is having difficulties.
Logfile Cannot Be Created
This error result from selection of an output folder to which write access is not available. This error typical occurs when using output paths on network shared drives or when the drive becomes full. If the error occurs check that the shared drive is still accessible and writable and/or that the output drive is not full.
Failed To Write To Logfile
This error occurs when Auto PeakTrace RP is not able to write the results to the current logfile. This can occur when:
- The shared drive where the output folder is located has become disconnected.
- The drive is full.
- The logfile is open by another program. This is the most common reason for this error,
Temporary Logfile Cannot be Found
This error occurs when the temporary logfile generated by peaktracerp.exe cannot be found or opened by Auto PeakTrace RP. This error occurs if the hard drive is full or if peaktracerp.exe does not have permission to write to the user temporary data folder. Check that the user running Auto PeakTrace RP has permission to write to User Documents folder.
Unicode Logfile Warning
Auto PeakTrace RP can now process traces within folder having Unicode (non-English) characters. Due to the way character encoding works on computers, it is not possible to print paths containing Unicode characters in ASCII (plain text files). Attempting to print Unicode characters in a plain text file so results in a “?” being substituted for any non-ASCII (non-English) character (e.g. \out\???? ?????? hello\ABI_3100_POP7_BD1_80cm_B.ab).
To avoid this issue Auto PeakTrace RP can now generate logfiles in Unicode format. Since this format may cause issues for downstream applications parsing the logfile, it is recommended that the Unicode logfile option is not used unless needed. The need for Unicode logfiles can be avoided by choosing input and output folders (or sub-folders) with only English characters. If your input or output folder paths contain Unicode characters, Auto PeakTrace will warn you with the following dialog window.
You can choose to ignore this warning and Auto PeakTrace RP will output all Unicode characters as “?”.
Auto PeakTrace RP License Warning
If your license is about to expire you should be provided with the following warnings. To obtain a new license please contact Nucleics or your local supplier.
Auto PeakTrace RP License Expired
If you obtain this error please contact Nucleics or your local supplier.
Only One Copy Open at a Time
You can only run copy of Auto PeakTrace RP per computer. You can share the same license code on different computers. This may cause the units report to be inaccurate until at least one trace is processed on a particular computer, but there is no issue is using as many copies of Auto PeakTrace (on different computers) with the same license.
Other Errors
The cause of this error is usually permission problem. Make sure that the user logged in has administrator rights.
The above two errors can be caused if files have been moved manually from the Auto PeakTrace RP program folder. To fix re-install Auto PeakTrace RP again.
What sequencers and run conditions are supported?
PeakTrace does not support all possible DNA sequencer run conditions. PeakTrace may not support some modified run conditions, or if you have used an unusual combinations of run voltage, polymer type, instrument, dye chemistry, and/or capillary lengths. If you find that your run condition is not supported please contact Nucleics as we can add support for new run conditions quickly once we have some example trace files. The following run conditions are currently supported:
ABI 3730, 3730xl, 3100 and 3130 Run Conditions
• POP7, BigDye3, 8.5kV, 50cm array traces
• POP7, BigDye1, 8.5kV, 50cm array traces
• POP7, BigDye3, 8.5kV, 36cm array traces
• POP7, BigDye1, 8.5kV, 36cm array traces
• POP7, BigDye3, 6.1kV (long run), 50cm array traces
• POP7, BigDye1, 6.1kV (long run), 50cm array traces
• POP7, BigDye3, >12kV (rapid run), 50cm array traces
• POP7, BigDye1, >12kV (rapid run), 50cm array traces
• POP7, BigDye3, >12kV (rapid run), 36cm array traces
• POP7, BigDye1, >12kV (rapid run), 36cm array traces
• POP7, ET Terminator, 8.5kV, 50cm array traces
• POP7, ET Terminator, 8.5kV, 36cm array traces
• POP7, BigDye3, 8.5kV, 80cm array traces
• POP7, BigDye1, 8.5kV, 80cm array traces
• POP6, BigDye3, 8.5kV, 80cm array traces
• POP6, BigDye3, 8.5kV, 50cm array traces
• POP6, BigDye1, 8.5kV, 50cm array traces
• POP6, BigDye3, 8.5kV, 36cm array traces
• POP6, BigDye1, 8.5kV, 36cm array traces
• POP6, ET Terminator, 8.5kV, 50cm array traces
• POP6, BigDye3, >12kV (rapid run), 50cm array traces
• POP6, BigDye1, >12kV (rapid run), 50cm array traces
• POP6, BigDye3, >12kV (rapid run), 36cm array traces
• POP6, BigDye1, >12kV (rapid run), 36cm array traces
• POP4, BigDye3, 8.5kV, 80cm array traces
• POP4, BigDye1, 8.5kV, 80cm array traces
• POP4, BigDye3, 8.5kV, 50cm array traces
• POP4, BigDye1, 8.5kV, 50cm array traces
• POP4, BigDye3, 8.5kV, 36cm array traces
• POP4, BigDye3, 8.5kV, 36cm array traces
• POP4, BigDye1, 8.5kV, 36cm array traces
• POP4, BigDye3, >12kV (rapid run), 36cm array traces
ABI 3500 Run Conditions
• POP7, BigDye3, 8.5kV, 50cm array traces
• POP7, BigDye1, 8.5kV, 50cm array traces
• POP7, BigDye3, >13kV (rapid run), 50cm array traces
• POP7, BigDye1, >13kV (rapid run), 50cm array traces
ABI 3700 Run Conditions
• POP5, BigDye3, 50cm array traces
• POP5, BigDye1, 50cm array traces
• POP6, BigDye3, 50cm array traces
• POP6, BigDye1, 50cm array traces
ABI 310 Run Conditions
• POP6, BigDye3, 50cm array traces
• POP6, BigDye1, 50cm array traces
• POP6, BigDye3, 36cm array traces
• POP6, BigDye1, 36cm array traces
• POP4, BigDye3, 36cm array traces
• POP6, BigDye3, 30cm array traces
• POP6, BigDye1, 30cm array traces
• POP4, BigDye3, 30cm array traces
• POP4, BigDye1, 30cm array traces
ABI 377 Run Conditions
• BigDye3 traces
• BigDye1 traces
• ET Terminator traces
MegaBACE Run Conditions
• BigDye3 traces
• ET Terminator traces
• ET Primer traces
MJ Research BioStation Run Conditions
• Dye Rhodamine traces
Why are my traces not being improved?
This is a complex question to answer as there can be many causes. The most common reason for PeakTrace not being able to improve the basecall is that your traces are lacking peak signal at the 3′ end of the trace. If the peak signal reaches close to the level of the detector noise then there is no real signal that can be basecalled.
Nucleics has developed a troubleshooting guide for automated DNA sequencing reactions to help researchers who are having difficulties with the quality of their DNA sequencing reactions. Included in this guide are protocol tips to help improve the quality and lower the cost of automated DNA sequencing. To ask question about any of these suggestions please visit the Nucleics DNA sequencing discussion forum.
How can I tell how many units I have left?
The number of units left on your license are shown at the bottom of the Options window.
If the number of units remain is 0 then you will need to purchase additional units. You should received a warning from Auto PeakTrace RP when you reach 100 units.
What option settings should I use?
The default setting are a good place to start.
It is also a good idea to select a small set of files (4 to 10) that span the typical range of trace quality you have and modify one setting at a time. Examine the traces after each change to see if it is helping on not.
DO NOT CHANGE SETTINGS WITHOUT KNOWING WHAT THEY DO!
Read what the various Options do. Avoid modifying the following options unless you have a good reason to do so.
- good quality threshold
- signal start peak
- clear range trim
- force processing
- trim improved traces only
- stealth mode
- noise raw data
If you are still having problems deciding what Option settings to use then we are happy to provide advice.
Help. I installed an update and my settings are blank!
Auto PeakTrace RP may delete your setting file when updating. This is to ensure that you are always using the correct settings. If you have not recorded your settings before updating you can refer to the text version which is saved in the Auto PeakTrace RP Application Data folder of the current user in the file called settings.txt. This file is only created once Auto PeakTrace RP is quit for the first time.
The settings.txt file can easily be accessed from the Start Menu via the Previous Settings link in the Auto PeakTrace RP folder.
Hints & Suggestions
Increase the DNA sequencing instrument data collection times
The optimal number of base peaks collected by the DNA sequencer is between 1100 and 1300 depending on the capillary length. If you are finding that PeakTrace is providing perfect basecalls right to the last base of your traces then you may want to modify your run condition to collect more data. You should not need to collect a lot more data, normally 5 to 10 minutes is sufficient to obtain the best from PeakTrace.
Lower the good base improvement threshold
If you are finding that many of your traces are not being improved by Auto PeakTrace RP and resulting in trace readlength improvement below cutoff, you may want to try using a lower or even negative good base improvement value. This will allow PeakTrace to process more traces at the risk of reducing the readlength of some traces. Use negative values with care!
Make use of the trace trim functions
The trim trace threshold can avoid many PeakTrace basecalling pitfalls by removing low quality regions. Try using a range of trim settings a values. A good way to do this is pick a couple of traces that have low quality regions and adjust the trim values to remove these regions while keeping the good quality regions. We think the quality score trim is the most useful. Take care with the “dangerous” trim settings.
Use the skip short trace function
This can avoid processing short traces (typically from short PCR products). These traces can rarely be improved by PeakTrace since KB has basecalled them to the maximum extent possible. Using this setting will save you the expense of processing traces that can’t be improved by PeakTrace.
Don’t modify option settings without knowing what they do
Auto PeakTrace RP allows you great control of most aspects of the PeakTrace processing and basecalling. Many of the settings only require modification under certain specific circumstances and are best left at their default values. The settings that should be modified with caution are:
- good quality threshold
- signal start peak
- clear range trim
- force processing
- trim improved traces only
- stealth mode
- noise raw data
Consider using the true profile setting
The true profile setting better reflects the underlying data signal strength and can be useful in visualizing the problem of rapid signal decline. This can give you and your end users valuable feedback on problems that are occurring in production.
Care when using custom run modules
When using custom run modules, the KB basecaller may not be able to basecall accurately. While this does not affect PeakTrace, if the fuse trace or fuse basecall options are used the poor quality of the KB basecalling will be carried through to the PeakTrace output file. It is recommended that if you are using a custom run module the you examine the KB output traces for accurate KB basecalling before using either the fuse trace or fuse basecall options.