Identifying mixed template sequencing traces
- The sequencing trace has two or more sequence peaks at the same location (Figure 1). The secondary peaks may be the same height as the primary peaks down to about 20%. Lower levels of mixed signal (below 20%) are normally base called well by the KB.
- The trace signal strength is good (>200U) in the raw channel.
- The quality scores are low with often less than 100 Q20+ bases.
- The base called sequence does not match the expected sequence or any known sequence in GenBank.
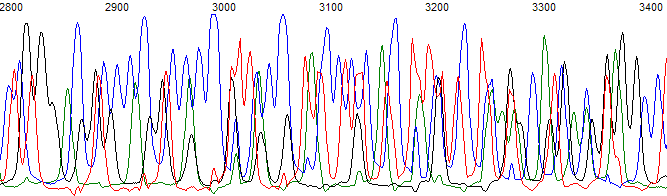
A
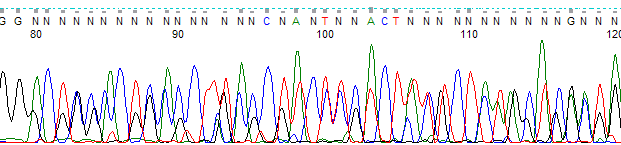
Example of a trace with mixed template. (A) Raw channel data. (B) Processed data channel at the same base location.
Causes of mixed template sequencing traces
- Two or more templates were present in the reaction. This is the most common cause of mixed template.
- A “double pick” of two colonies. This can when the colonies are too close together on the colony plate.
- Two primers were present in the sequencing reactions. This can occur when using premixed PCR regents for sequencing where the primer stock is actually a mix of universal forward and reverse oligonucleotide primers.
- The PCR fragment was not purified of leftover primers before sequencing.
- Two priming sites are present in DNA template. This can occur when a PCR product with universal priming site tails is cloned into a plasmid.
- Poor quality PCR template containing multiple DNA fragments was used.
- Too low a primer annealing temperature was used in the sequencing reaction.
- Different sequencing reactions were accidentally mixed at the clean up stage. This can also sometime occur if the same tip is used with out rinsing.
Solving mixed template sequencing problems
- Makes sure only one DNA template is present. Prepare a new plasmid prep making sure that only one colony is selected. If sequencing a PCR product check that only one PCR product is present by running an agarose gel. Remember that that even a relatively low amount of a small PCR product can cause mixed template problems.
- Check the template for possible multiple priming sites. If two sites are present use a different primer. This can often occur when a fragment contain the priming is sub-cloned into a vector that also contains the priming site. Be very careful of this problem when using the M13 universal primers.
- Insure that you use a PCR clean-up protocol that remove leftover PCR primers. Even low levels of the PCR primers can cause mixed signal problems, especially if they have a high annealing temperature.
- Check the predicted melting temperature of the sequencing primer. If it is more than 5˚C above the annealing temperature used in the sequencing reaction then raise the annealing temperature. Because of the inclusion dITP in the BigDye sequencing mix the annealing/extension temperature can’t be raised above 60˚C. If your primer is still misannealing at 60˚C then synthesize a new primer (this is easily done by removing bases from the 5′ end until the Tm is below 60˚C).
For more information on automated QC tracking of sequencing traces please visit the QualTrace DNA sequencing analysis software page.
Return to the main DNA sequencing troubleshooting page.
