Identification of Micro-satellite Slippage
- The trace becomes mixed after a long dinucleotide (alternating two bases) run (as seen in Figure 1.). Can also occur for trinucleotide (repeating of 3 base sequence) runs.
- Looks similar to the effects of mononucleotide slippage, however it is caused by different mechanisms.
- The cause is at the template preparation stage when polymerase chain reaction (PCR) amplification is used.
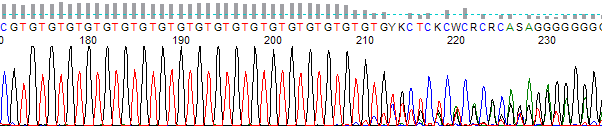
Figure 1. Dinucleotide G and T run with slippage occurring.
Figure 2 shows an example of trinucleotide run which did not result in slippage. This is due to the C and G having stronger linkage, which helps to prevent misalignment or looping of the DNA.
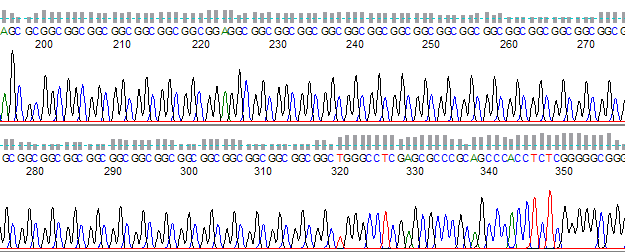
Figure 2. Trinucleotide repeat of GGC without slippage.
Causes of Microsatellite Slippage
- During PCR amplification of the template the repeated bases “breathe” and misalign (loop out) creating some fragments to be a few repeats shorter or longer.
- The length of the microsatellite has a directly proportional effect on the extend of the slippage that occurs (i.e. the more repeats, the worse the problem).
Solving Microsatellite Slippage Problems
- Sequence the DNA template from both directions. For example, if the template shows slippage after using the forward primer then sequence the template using the reverse primer. You may need to make a custom primers to do this if the template is large.
- Use a custom primer designed to hybridize just outside the dinucleotide run region.
- Use the sequence-by-mutagenesis (SAM) approach to avoid have long dinucleotide runs in your templates.
Return to the main DNA sequencing troubleshooting page.