One of the new features of PeakTrace 6.20 is user adjustment of the PCR trim base. In previous releases of PeakTrace the PCR trim base was set automatically using the skip short base setting value. Since skip short base could not be set to higher than 500, it meant that the PeakTrace basecalling of large PCR products (i.e. those larger than 500 bases) could result in traces with additional bases after the PCR product if there was low level template contamination and/or signal bleed through from an adjacent capillary present. This effect is shown below.
Figure 1 shows a KB basecalled trace of a ~530 base-pair PCR fragment. The KB Basecaller™ ends the basecalling at 3 A bases corresponding to the end of the PCR fragment sequence.
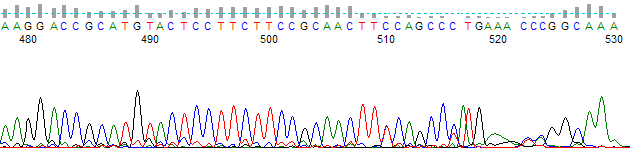
Figure 1. KB Basecalled Trace
Figure 2 shows the same trace as in Figure 1, but basecalled using PeakTrace and the default skip short/PCR trim base of 500. Due to low-level template contamination the sequencing continues past the last 3 A bases of the PCR fragment. This extra trace signal is real, but is not wanted by the end user.
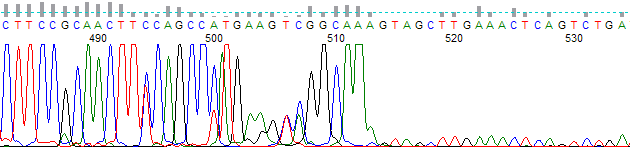
Figure 2. PeakTrace Basecalling with a PCR Trim Base of 500
Figure 3 shows the same trace as in Figures 1 & 2, but basecalled using PeakTrace and a skip short/PCR trim base of 700. The trace is now trimmed to the last 3 A bases of the PCR fragment (as in Figure 1), thus removing the contamination sequence.
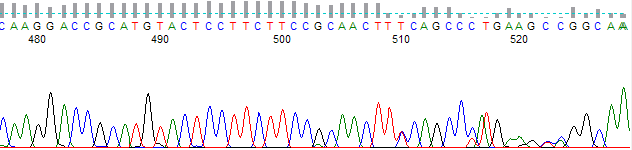
Figure 3. PeakTrace Basecalling with a PCR Trim Base of 700
Pitfalls with using skip short/PCR trim
- Setting the skip short/PCR trim base to any value greater than 500 will turn off the skip short traces option, resulting in more traces being basecalled by PeakTrace (this may or may not be a desirable outcome).
- Setting the skip short/PCR trim base to a high value (i.e over 850) may result in weak signal traces being falsely detected as PCR product traces resulting in premature trimming.
For these two reasons it is recommended that the skip short/PCR trim base only be adjusted when sequencing large PCR fragments larger than 500 bases.