One of the new features introduced in PeakTrace 6.40 was Set ABI Limits. This option changes the PeakTrace trimming functions from hard trims (i.e. actual removal of the low quality regions) to the same soft (virtual) trimming that is performed by the KB Basecaller’s Post-Processor module. If trace trimming is selected within Sequencing Analysis, then KB will remove the low quality regions from the trace by setting the ABI Limits values in the trace file to indicate the high quality region. When the trace or basecall is viewed in Sequencing Analysis or Sequence Scanner, the trimmed basecalls are still present, but the peaks and bases are now grey (Figure 1).
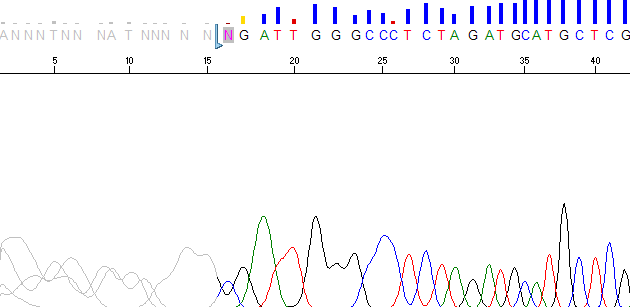
Figure 1. Example trace viewed in Sequence Scanner with the ABI limits range set. The ABI limits region starts at base 15.
PeakTrace is now also able to set the ABI Limits values by using the Advanced Option Set ABI Limits with trace trimming. Instead of the hard removal of the low quality regions, these regions are “greyed out” when viewed in Sequence Scanner or Sequencing Analysis – just like KB (Figures 2 & 3).
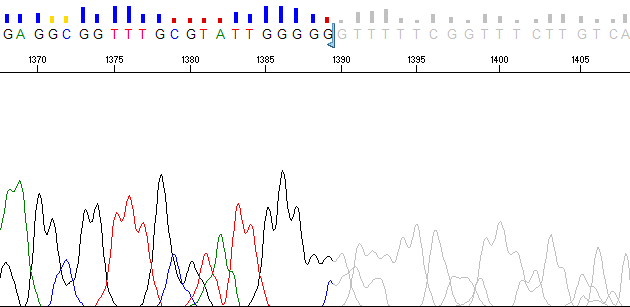
Figure 2. PeakTrace basecalled trace using Set ABI Limits and Q Average Trim of 12 & 20. The last low quality region has been greyed out.

Figure 3. PeakTrace basecall using Set ABI Limits and Q Average Trim of 12 & 20. The last low quality bases are shown in grey and the high quality regions in blue.
One of the limitations with using Set ABI Limits is that none of the non-ABI trace viewers are able to use the ABI limits region. For example, Figure 4 shows the same trace as shown in Figure 2, but in FinchTV. The low quality region after base 1390 is not shown greyed out or trimmed by FinchTV.
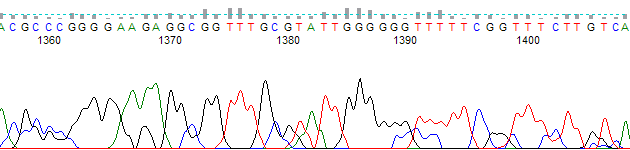
Set ABI Limits is a very useful addition for those users of PeakTrace who wish to replicate the results provided by the KB Basecaller as close as possible. It provides the same “greyed out” trimming of the low quality regions that Sequencing Analysis Post-Processing provides.
Users of this setting are encouraged to upgrade to PeakTrace 6.42 or greater as this release has introduced a workaround for a bug in Sequencing Analysis that stops the display of the 3’ trimmed region when the 5’ end is untrimmed.