Troubleshooting DNA Sequencing Problems
Automated DNA sequencing is one of the most common and normally most robust techniques performed in molecular biological laboratories. Unfortunately, it does not always work and when it doesn’t it can be very difficult to work out what went wrong. Fortunately, most failed (or sub-optimal) DNA sequencing results have only a limited number of causes. To help the troubleshooting of DNA sequencing problems we have created a series of guides for identifying the most common causes. These guides also includes tips on how to overcome each problem type, along with more general tips for improving DNA sequencing quality.
If you have any comments or questions about these guides then please contact our support team at {This email is obscured. Your must have javascript enabled to see it} as we would love to hear your suggestions.
How To Identify The Causes of DNA Sequencing Failures
Identifying the cause of a poor DNA sequencing result can often be very difficult as a particular sequencing problem may have many different causes, or be the result of multiple interacting factors. Often the only way to work out the real cause of a particular problem is to perform a process of elimination.
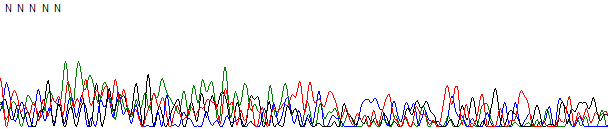
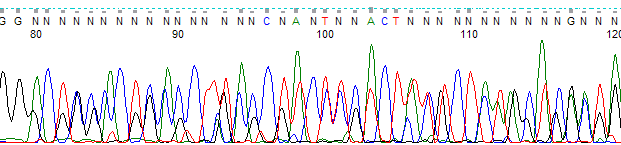
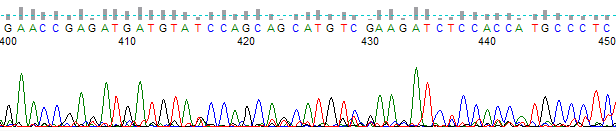
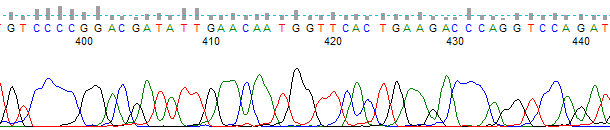
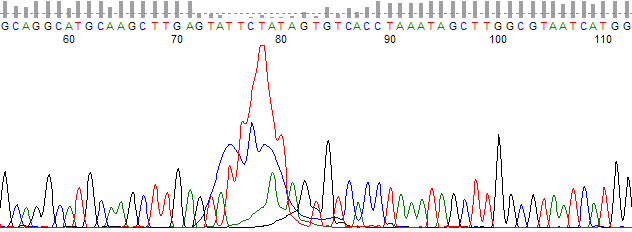
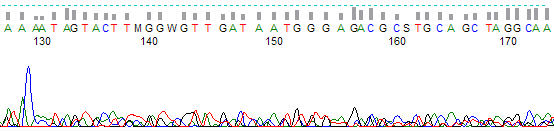
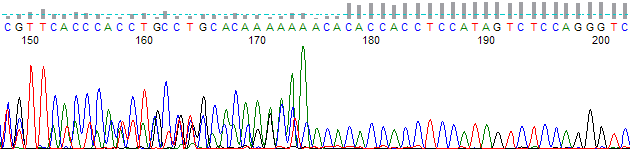
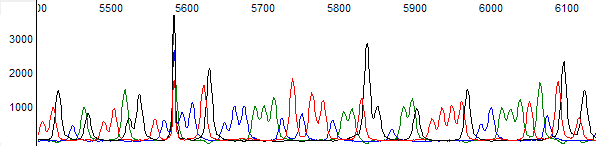
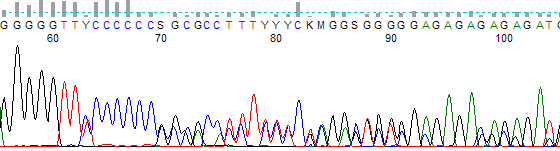
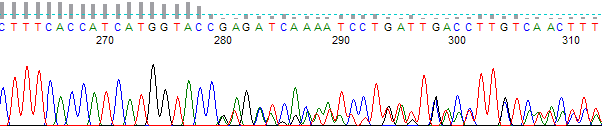
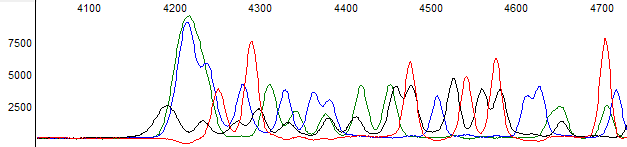
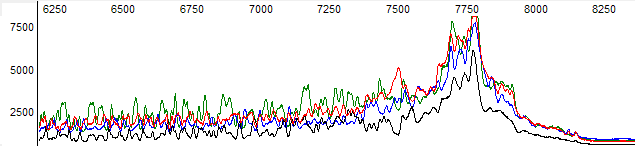
This process can be greatly simplified by visually examining both the raw and processed data chromatograms of the sequencing traces. In the following guide we have provided detailed information on the most commonly observed sequencing problems, along with suggestions on their most likely causes. The causes are listed in order from the most common to least common. We have also included solutions (where known) on how to overcome each sequencing problem type.
An alternative to manual inspection (which becomes very labor intensive if you are running more than a few traces) is to use an automated trace analysis system like our QualTrace III DNA sequencing QC software. QualTrace III will automatically scan the traces for many different sequencing problems, and because it works by analysing the raw data, it is able to be used with any basecaller.
To see how QualTrace III can help in DNA sequencing troubleshooting we have created a free, online version of QualTrace III where you can upload your own traces and have QualTrace III analyze them for any problems.
The Most Common Automated DNA Sequencing Problems
The following guides give examples of the major causes of Sanger DNA sequencing problems, describes how to identify them, and how to solve the underlying problems. More details are provided on each problem by clicking on the title.
Failed DNA Sequencing Reactions →
Mixed Trace Signal (multiple peaks) →
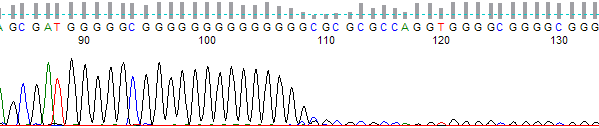
Short Read Lengths or Poor Quality Basecalls →
Poorly Resolved Trace Peaks (blurry peaks) →
Excess Free Dye (“dye blobs” peaks) →
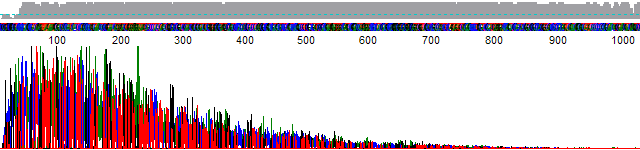
Weak or “Noisy” Trace Peaks →
PCR or Primer Dimer Formation in the Sequencing Reaction →
Sharp Signal Spikes in the Trace →
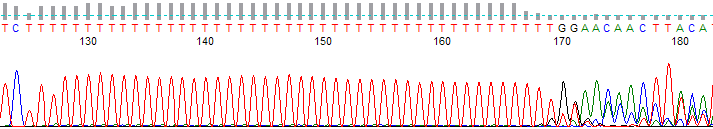
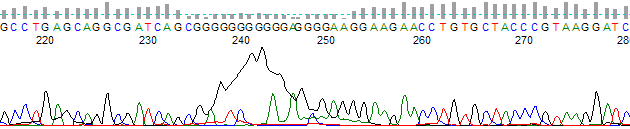
Dinucleotide Run (Microsatellite) Slippage →
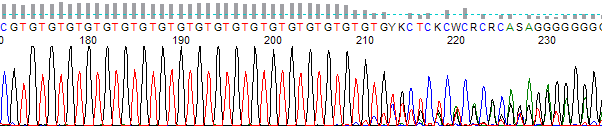
Dinucleotide Run (Microsatillite) Slippage